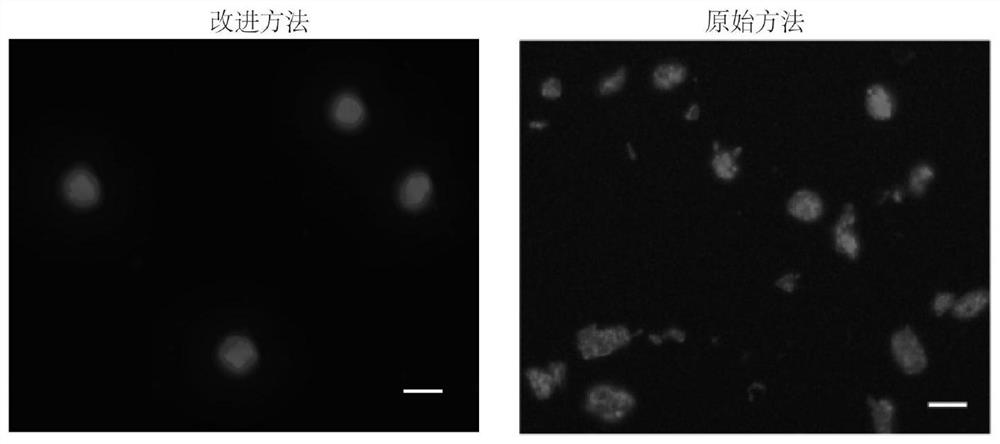
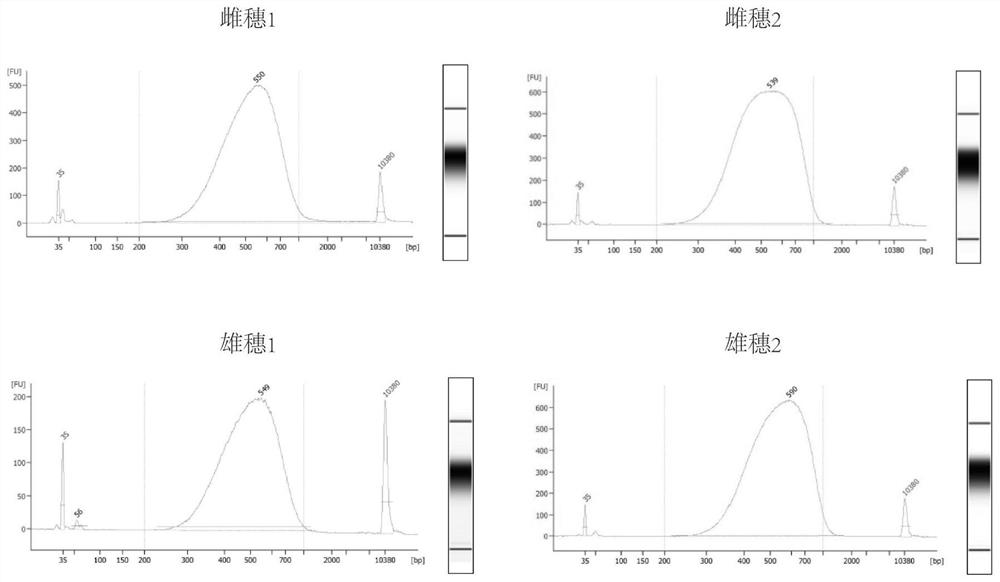
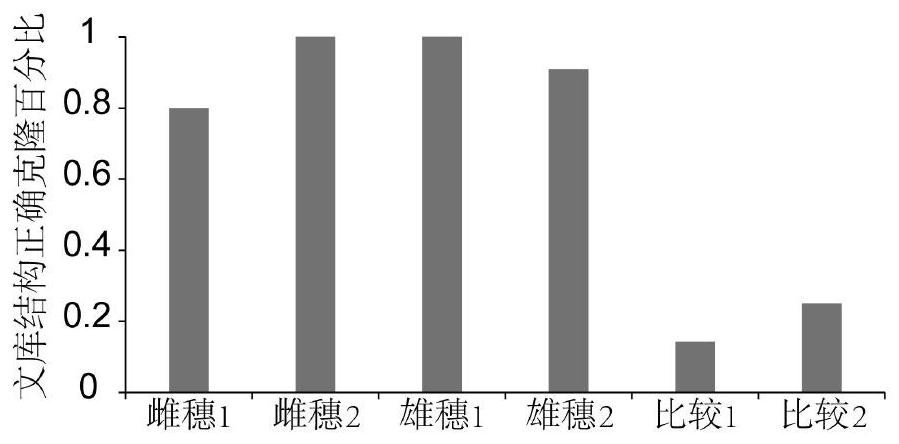
A simple and effective method for constructing plant long-fragment in situ DLO Hi-C sequencing library
A technology of sequencing library and construction method, which is applied in the field of construction of plant long-fragment insituDLOHi-C sequencing library, can solve the problems of sequencing cost and resolution limitation, and achieves the improvement of genome alignment rate, good integrity, and avoidance of completeness. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0059] The preparation method of the linker comprises: after the pre-sense strand and the post-sense strand are hybridized to obtain the linker, the nucleotide sequence of the pre-sense strand is shown in SEQ ID No.1, and the nucleotide sequence of the post-sense strand is The sequence is shown as SEQ ID No.2;
[0060] 6) centrifuging and rinsing the linker connector obtained in step 5) to obtain a cell nucleus, mixing the cell nucleus with the phosphorylation system at the end of the linker, and bathing in water at 37° C. for 25 to 35 minutes to obtain a second water bath;
[0061] The linker terminal phosphorylation system component is 170μl ddH 2 O, 20 μl of 10×T4 ligase buffer, 10 μl of T4 polynucleotide kinase with enzyme activity of 10 unit / μl and 5 μl of Triton X-100 solution with a concentration of 20% by volume;
[0062] 7) After mixing the second water bath in step 6) with the connection system, at 20-25°C, connect the chromatin in the nucleus for 1.5-3 hours to obt...
Embodiment 1
[0101] 1. Take freshly harvested 2-4mm long tassels and female ears of maize at the development stage as tissue samples, carry out 2 biological repetitions respectively, and totally 4 complete experiments, named as tassel 1, tassel 2, Ear 1 and ear 2, and the same experimental procedures were used in the four experiments to verify the stability and reliability of the method.
[0102] 2. Put the tissue in an EP tube of appropriate size, add formaldehyde cross-linking buffer (1×PBS containing 1% sigma formaldehyde), fix under vacuum for 15 min for cross-linking, and add glycine to a concentration of 0.2M to stop cross-linking. Immediately afterwards, the plant tissue was rinsed three times with double distilled water, and directly proceeded to the next step or frozen at -80°C for storage.
[0103] 3. Take 0.1-2g of the cross-linked tissue sample and place it in a plate containing PP buffer (1×PBS containing protease inhibitors) pre-cooled on ice, and mash the tissue as much as p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com