A microfluidic chip and its dna calculation method for solving the minimum set covering problem
A technology for microfluidic chips and covering problems, applied in fluid controllers, chemical instruments and methods, and laboratory containers, etc., can solve problems such as complex and large drive systems, difficulty in miniaturization, and complex processes, and achieve the goal of overcoming series problems. Large amount of calculation, improved reliability, and simple chip structure
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
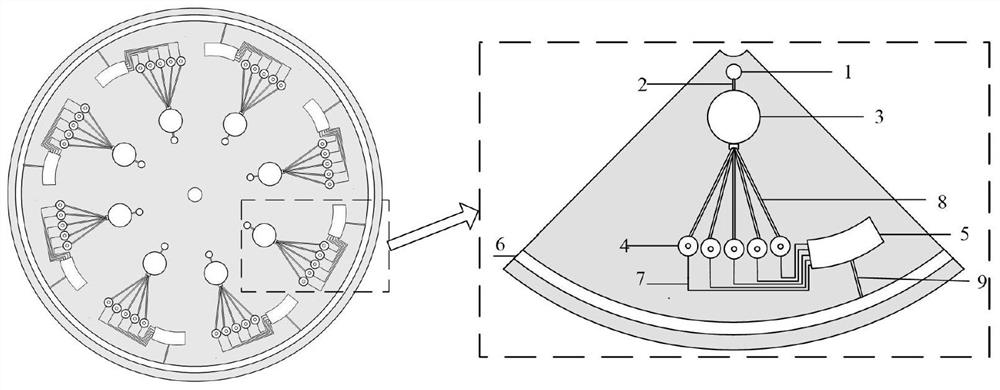
[0039] with S={S 1 ,S 2 ,S 3 ,S 4 ,S 5}, C = {C 1 ,C 2 ,C 3 ,C 4}, C 1 ={S 1 ,S 3 ,S 4 ,S 5}, C 2 ={S 1 ,S 4}, C 3 ={S 2 ,S 5}, C 4 ={S 2 ,S 3 ,S 4} as an example, since the set C has 4 elements, which requires 16 repeating units, two pieces such as figure 1 The chip shown to solve the MSC problem, the specific process is as follows:
[0040] (1) Design the chip and make a number mark, the design is as follows Figure 4 The chip shown is 2 slices, and the 16 units are sequentially placed in accordance with the C 1 ,C 2 ,...C 4 The values in different combinations are numbered and marked, as shown in Table 3;
[0041] Table 3: Unit Numbers
[0042]
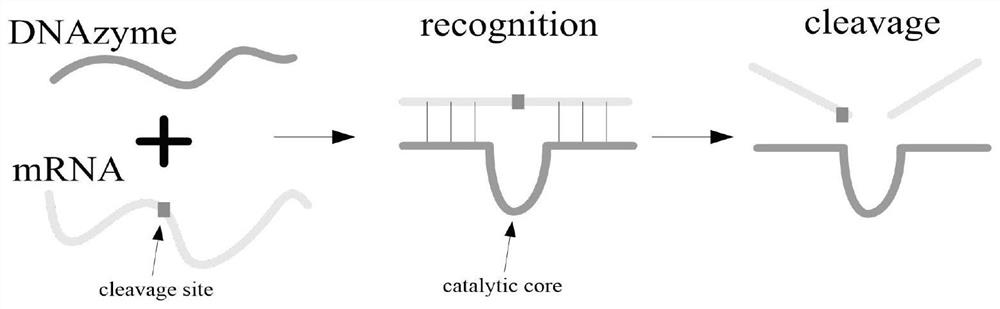
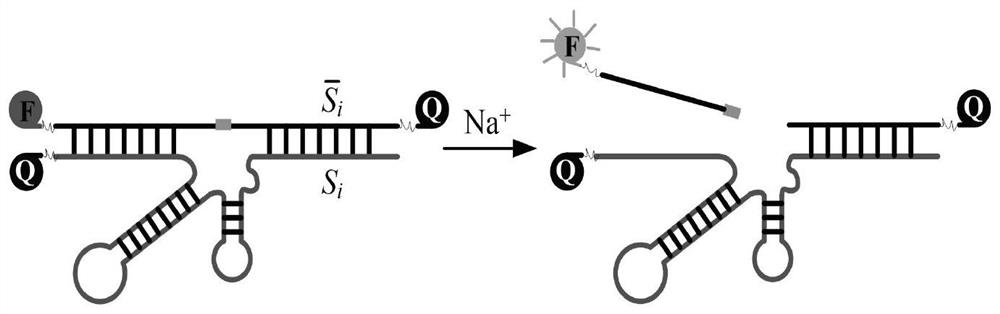
[0043] (2) Encoding the DNAzyme and RNA sequences required for synthesis, as shown in Table 4, add the mRNA sequence solution representing each element in the set S to the reaction pool d (a~e) of each repeating unit in sequence and the reaction The required Na+; then according to the numbering in ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com