3-sterone-1, 2-dehydrogenase as well as gene sequence and application thereof
A gene sequence and dehydrogenase technology, which is applied to 3-sterone-1,2-dehydrogenase and its gene sequence and application fields, can solve the problems of low enzyme activity and achieve the effects of high enzyme activity and increased solubility
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
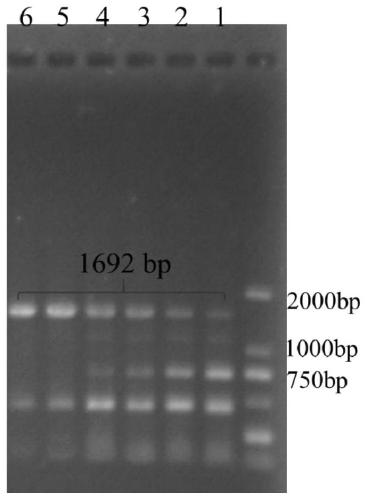
[0037] Embodiment 1: PCR amplification KsdD211 gene
[0038] From the Mycobacterium strain HGMS2GL genome, the KsdD211 target gene fragment was amplified by PCR with a size of 1692bp. The PCR primers were: the front primer KsdD211-F and the back primer KsdD211-R. ℃, 54℃, 56℃, 60℃. The results of running agarose gel analysis are as follows: figure 1 shown.
[0039] The PCR products at 56°C and 60°C were selected for enzyme digestion and ligation. The digestion reaction system is 50 μL: PCR product: 1ug; BamHI 2U; EcoRI 2U; 10×K buffer 5ul; H 2 O 1ul; 37°C, time: 3h. The reaction product was purified with a PCR purification kit (Tiangen Biotechnology, Beijing) and used for purification of the pRSV vector. The pRSV vector was digested and purified using the same method and system as above. After digestion, the vector pRSV and the target fragment KsdD211 were ligated by T4 ligase reaction, and the 20 μL reaction system was ligated overnight at 4°C. Table 1 shows the pRSV-Ks...
Embodiment 2
[0044] Example 2: KsdD211 gene sequencing
[0045] Sequencing was carried out at Wuhan Boshang Sequencing Company, and the analysis of the sequencing results was correct. See SEQ ID NO.1 for the KsdD211 gene. The amino acid sequence of KsdD211 is shown in SEQ ID NO.2.
Embodiment 3
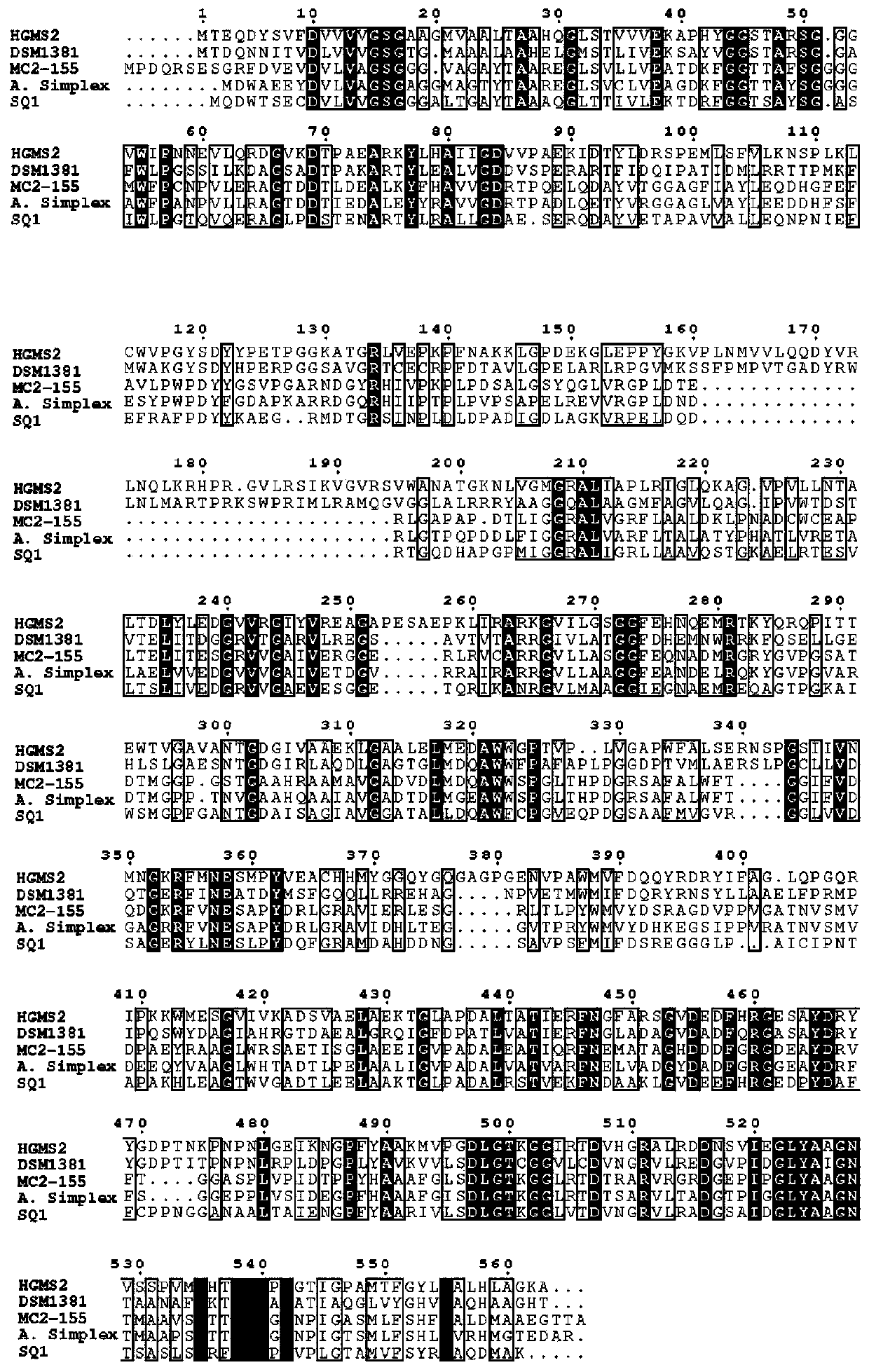
[0046] Example 3: Optimization of KsdD211 enzyme structure based on comparison of amino acid sequence and three-dimensional protein structure
[0047] Compared with other KsdD amino acid sequences, it is found that the amino acid sequence of KsdD211 is very different, especially in the active center, different KsdD enzymes have different biases, the main difference is that Leu136 and Val364 are hydrophobic amino acids ( image 3 ), which is beneficial to substrate binding, but unfavorable for dehydrogenation reaction. Therefore, changing to carry out amino acid, changing Leu136 to His, Thr, Ser, His, Glu or Asp amino acid is beneficial for dehydrogenation reaction. Similarly, the change of Val364 to His, Thr, Ser, His, Glu or Asp amino acid is also beneficial to the dehydrogenation reaction. The three-dimensional structure of KsdD211 shows that there are five redundant loop regions in the protein conformation, namely Asp161-S188, Pro253-Glu257, Arg340-Pro344, Gly377-G382 and A...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com