Method for extracting DNA of single chironomidae pupal stage ecdysis
A mosquito family, pupal stage technology, applied in DNA preparation, recombinant DNA technology, biochemical equipment and methods, etc., can solve the problem that the total DNA purity and quality cannot meet the requirements of subsequent experiments, and the molecular identification of chironomid pupa skin is difficult to carry out. Unable to extract pupa skin DNA and other problems to prevent interference with extraction
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
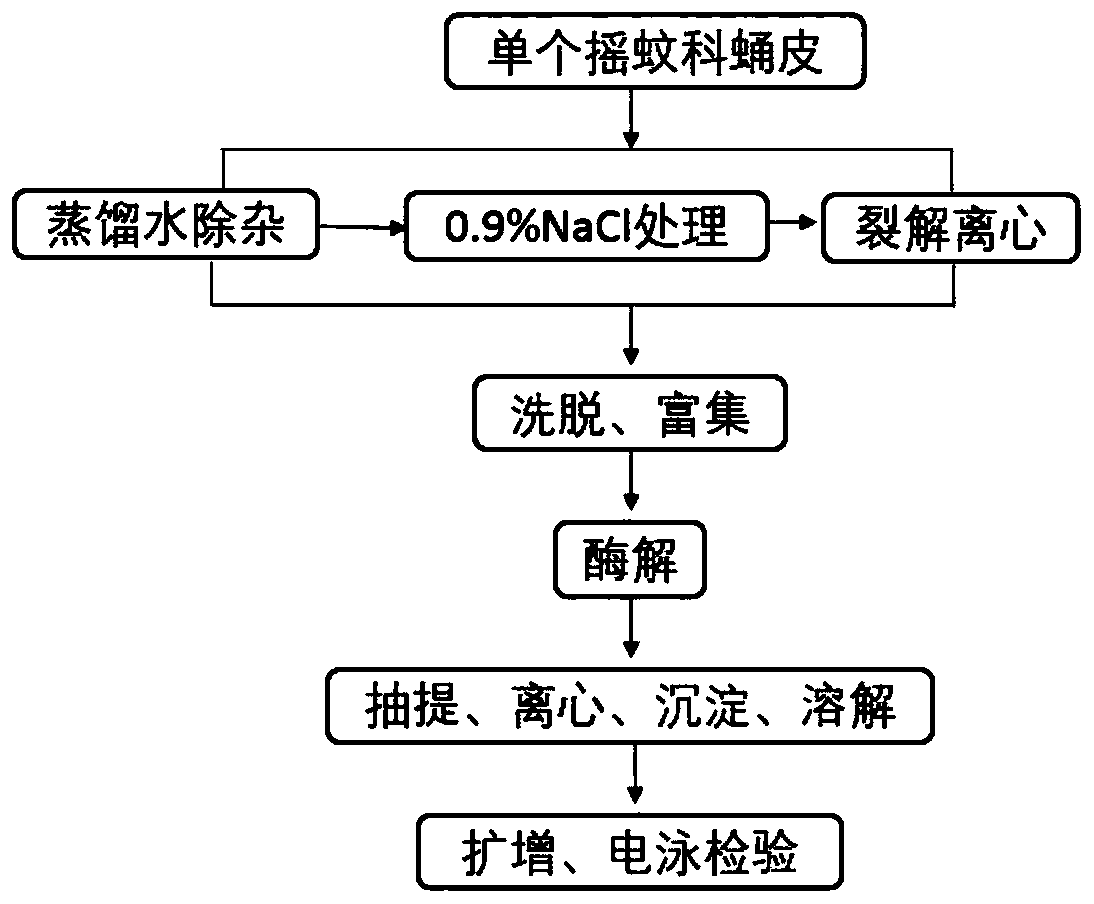
[0051] (1) Preprocessing:
[0052] (11) Take a single molt of chironomus pupal stage, soak it in distilled water at 25°C for 5 minutes, then replace the distilled water and repeat once. The amount of distilled water added is 1-2 cm above the molt of chironomus pupal stage to remove impurities and prevent interference with extraction. .
[0053] (12) After soaking in distilled water, soak in 0.9% NaCl solution again. The amount added is 1-2 cm above the molting stage of chironomus pupal stage. Change it every 1 hour, and change twice for a total of 3 hours. .
[0054] (2) Lysis centrifugation:
[0055] (21) Preparation of lysate: Weigh 2g of CTAB, 0.5g of SDS, 6ml of 0.5M EDTA, 10ml of 1M Tris-HCl, 20ml of 5M NaCl, 1g of PVP and 0.1ml of β-mercaptoethanol, and make up to 100ml with distilled water.
[0056] (22) Put the pretreated chironomus pupae molt in the Ep tube, use dissecting scissors to cut the chironomus pupae molt into small segments with a width less than 2mm, add 1...
Embodiment 2
[0069] Example 2 PCR Amplification of Mitochondrial Cytochrome C Oxidase Subunit I Gene (COI Gene)
[0070] (1) PCR reaction system:
[0071] Total system 50 μl: ddH 2 O 32.4μl, 10ⅹPCR Buffer 5μl, dNTPs (2.5mmol / L) 4μl, universal primer LCO and HCO (10μmol / L) 2μl each, Taq enzyme (500U, 2.5U / ul) 0.6μl, DNA template 4μl.
[0072] (2) PCR reaction conditions:
[0073] Pre-denaturation at 94°C for 5min; denaturation at 94°C for 10s; annealing at 55°C for 30s; extension at 72°C for 60s; 35 cycles; extension at 72°C for 10min; storage at 4°C.
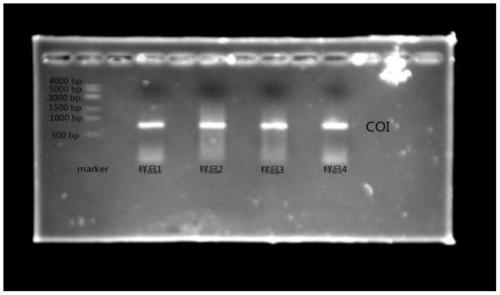
[0074] (3) Configure 1% agarose gel for electrophoresis detection, the results are as follows figure 2 shown.
[0075] figure 2 Among them, the first lane is 8Kp Marker, and the second, third, fourth, and fifth lanes are the PCR results after extracting DNA from Chironomidae pupal molt (Propsilocerus akamusi) of the same species, same size, and same eclosion time After four sets of repetitions, the method of the present invention can...
Embodiment 3
[0077] Embodiment 3 DNA extraction method comparative test
[0078] Verify the difference in the effect of the conventional method and the method described in Example 1 on the extraction of DNA from a single chironomid pupal stage molt.
[0079] According to the traditional CTAB method and the DNA extraction kit sold in the market, according to the steps and the method described in Example 1, take 1 μl and pass through the ddH 2 After O was dissolved, the DNA extracted according to the CTAB method, the kit method and the method described in Example 1 was detected using a NanoDrop 2000 instrument.
[0080] NanoDrop 2000 uses a high-energy xenon lamp. After the light passes through the test sample, part of the light is absorbed. NanoDrop decomposes the light with complex components after passing through the sample into spectral lines, and calculates the absorbance value of the sample to convert it into the concentration of the sample.
[0081] Calculation formula: A=-lg(I / I0)=-...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com