DNA molecular machine for detecting exosomes
A DNA molecule and exosome technology, which is applied in the fields of biotechnology and medical diagnosis, can solve the problems of specific response of DNA molecular machines and the limitation of autonomous energy supply, and achieves low cost, expanded detection range, high specificity and sensitivity Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Example 1: Cell culture and exosome collection
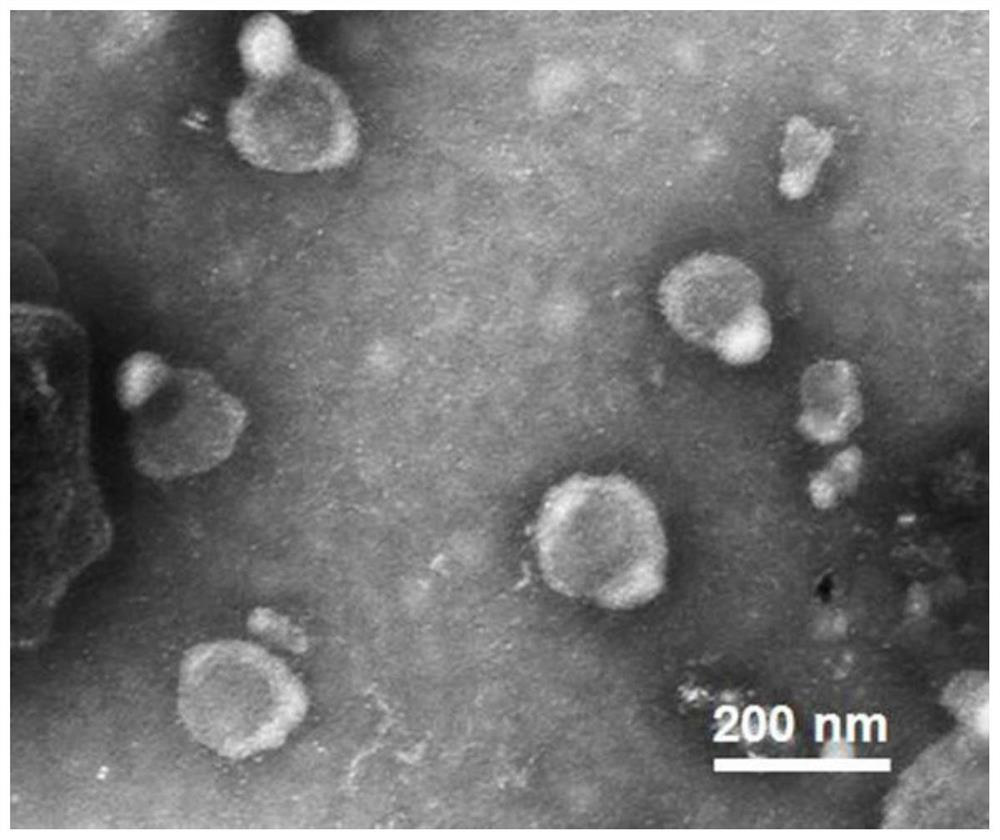
[0042] Cells such as MCF-7, HepG2 and HeLa need to be cultured with RPMI 1640 complete medium containing 10% fetal bovine serum, 1% penicillin and 100 μg / mL streptomycin, and then placed in a cell culture incubator (37°C humid environment, 5% CO 2) proliferate. When the cell coverage area reaches about 80%-90%, suck out the 1640 complete medium in the cell culture dish (150mm), wash twice with a small amount of sterile PBS (pH 7.4), and then add a certain amount of 1640 basal medium , and incubated in a cell culture incubator. After 48 hours, all the liquid in the culture dish was sucked out, and exosomes from various tumor cell lines were isolated from the 1640 basal medium by ultracentrifugation. Briefly, cells, dead cells, and cell clearings were centrifuged at 300 g, 4°C for 10 min, 1000 g, 4°C for 10 min, and 10,000 g, 4°C for 30 min to remove the precipitate, and the supernatant was collected. Afterwards, the su...
Embodiment 2
[0044] Embodiment 2: Construction of DNA molecular machine
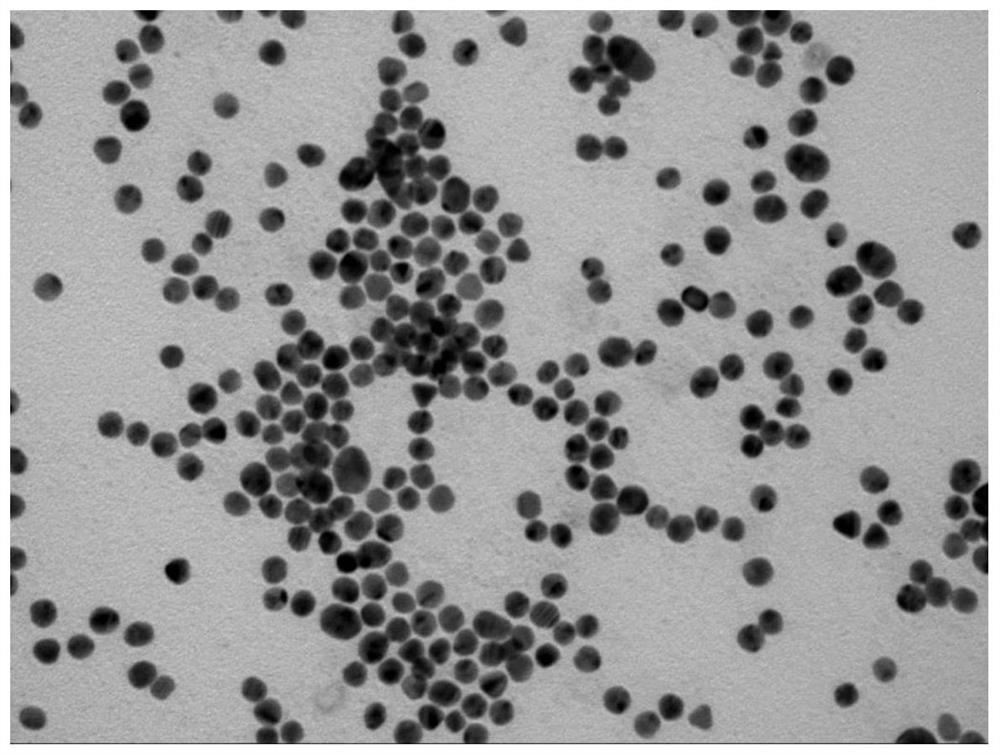
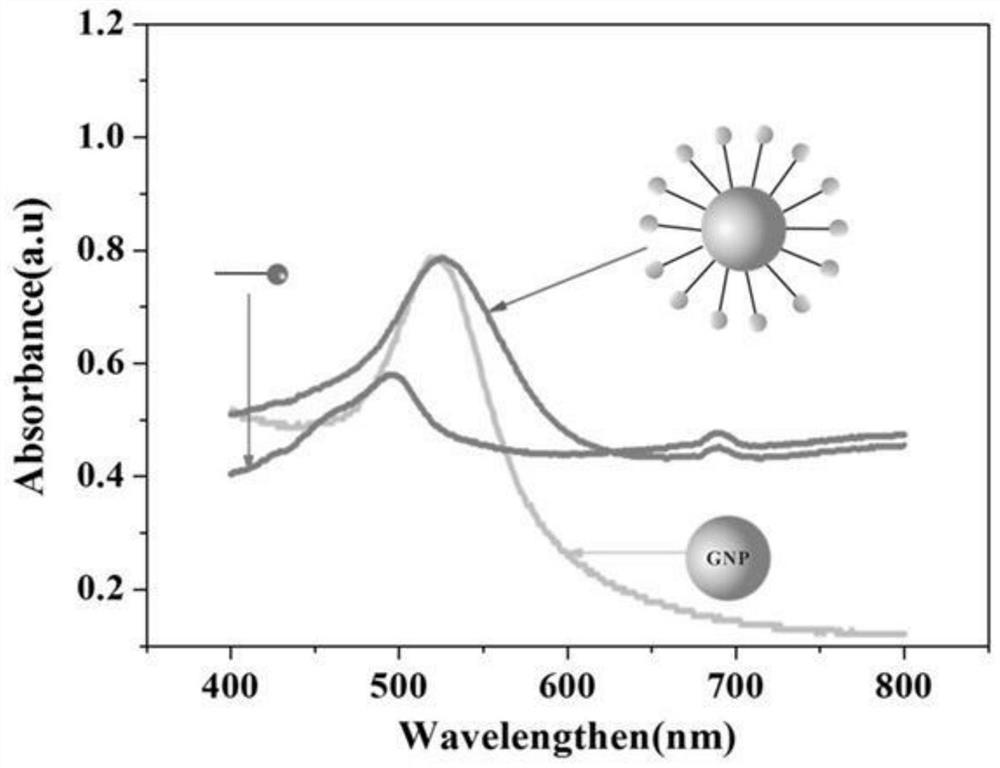
[0045] The DNA molecular machine is based on GNP with a diameter of 15nm, and the Substrate and the Motor chain locked by CD63 aptamer are connected to the surface of GNP through -SH at one end of the chain. To synthesize 15nm GNPs, 100mg HAuCl 4 ·3H 2 O was dissolved in 300 mL deionized water and the solution was heated to boiling. A solution of 264 mg of sodium citrate dissolved in 30 mL of water was quickly added to the mixture. The color of the solution changed from yellow to blue, and then to burgundy red within 5 minutes, and kept vigorously stirring at boiling temperature for 10 minutes, and the GNP solution was obtained after the solution was cooled to room temperature.
[0046] In order to completely lock the Motor chain, 1 μL Motor (1 mM) and 2 μL CD63 aptamer (1 mM) were mixed in 97 μL PBS (pH 7.4) for annealing. The mixture was heated to 95°C and gradually cooled to 4°C over 30 min. Then, 18 μL Subst...
Embodiment 3
[0049] Example 3: Determining the upload volume of Substrate on GNP
[0050] To determine the number of substrate strands per GNP, the conjugated Substrate was dissociated from the GNP surface by cleavage of Au-S using 2-mercaptoethanol. Briefly, DNA nanomachines (40 μL) were diluted to 200 μL with PBS and 2 μL 2-mercaptoethanol was added, placed on a shaker in the dark and shaken overnight, then the mixture was centrifuged and the supernatant containing the released substrate was measured by fluorescence spectrometer . According to the standard curve, the fluorescence intensity was converted to the molar concentration of the substrate chain. The average substrate number per particle was obtained by dividing the measured substrate molar concentration by the raw GNP concentration. In this way, it is estimated that the number of Substrates on the GNP is 136 per particle. The molar ratio of substrate chains to Motor chains in the conjugation reaction was 1:18. Therefore, abou...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Particle size | aaaaa | aaaaa |
| Particle size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com