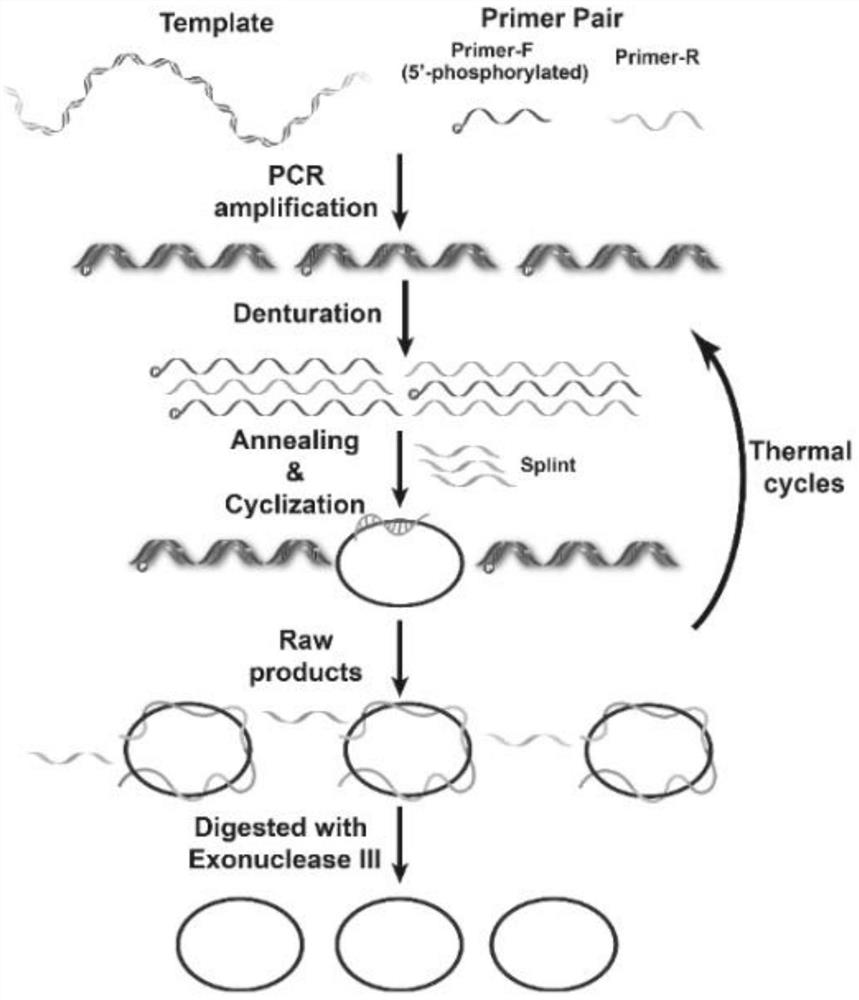
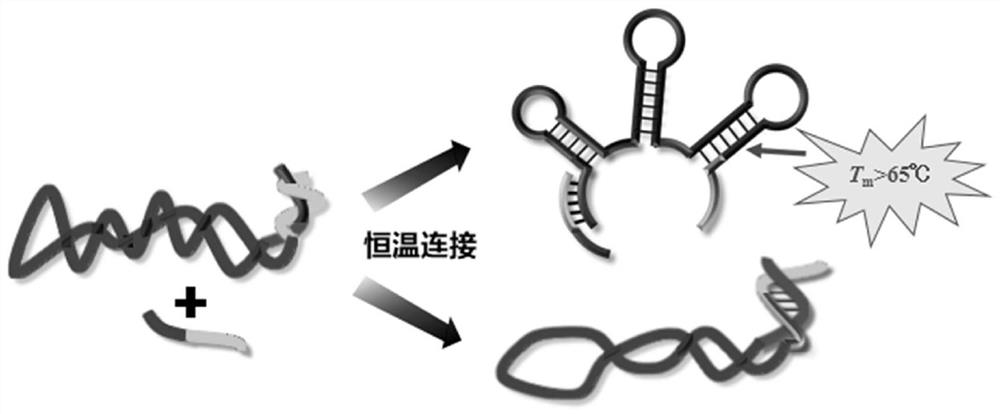
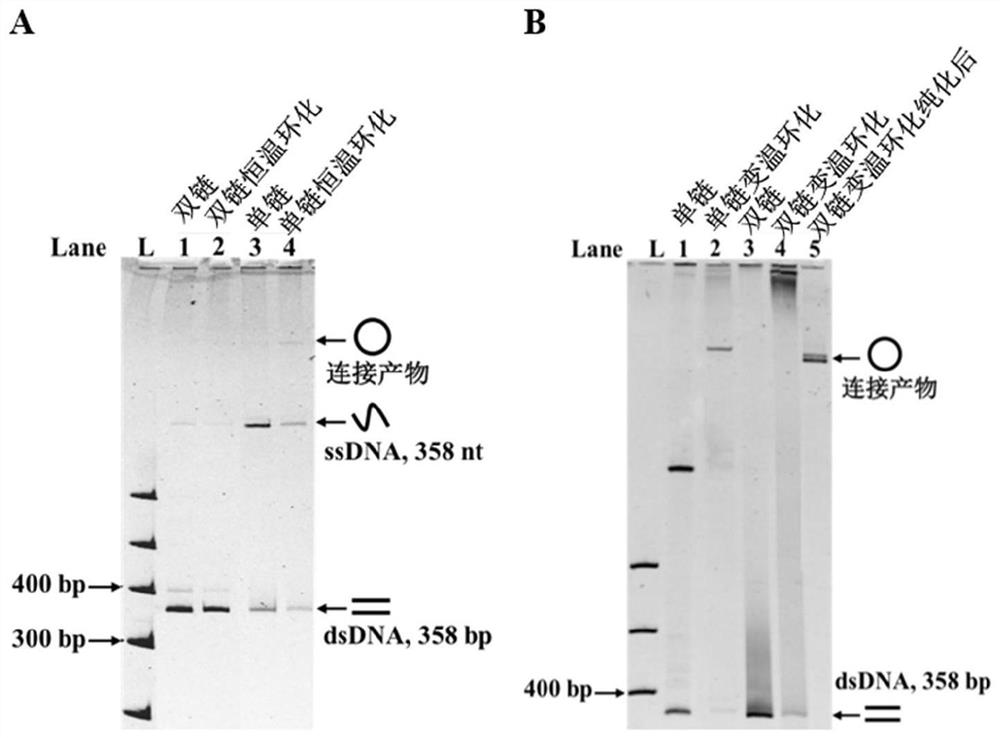
Preparation method of single-stranded circular DNA
A single-stranded, circular technology, applied in the field of nucleic acids, can solve the problems of low connection efficiency, difficult to open, difficult to prepare single-stranded DNA, etc., and achieve the effect of improving yield and wide application range.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] 1) Raw materials
[0035] aPCR primer chain F (358EX-F) (5'→3'): GTCTTGAGTCCAACCCGG (18nt in length, SEQ ID NO: 1)
[0036] aPCR primer chain R (358EX-R) (5'→3'): ATGACCAAAATCCCTTAACG (5'-phosphorylated, 20nt in length, SEQ ID NO: 2)
[0037] splint(5'→3'): CGTTAAGGGATTTTGGTCATGTCTTGAGTCCAACCCGGTA (40nt in length, SEQ ID NO: 3)
[0038] Template source: Escherichia coli plasmid pUC18 (Thermo Fisher Scientific Biotechnology Co., Ltd.)
[0039] 2) Preparation of medium-length single-stranded and double-stranded DNA
[0040] Asymmetric PCR (aPCR) was used to prepare 358nt single-stranded DNA, and PCR was used to prepare 358bp double-stranded DNA. The PCR system is: [pUC 18]=2×104copies / μL, [358EX-F]=0.2μM, [358EX-R]=0.2μM, [Phanta MaxMaster Mix]=1×, total volume 20μL; 95℃ pre-denaturation 5min, denaturation at 95°C for 15s, annealing at 55°C for 15s, extension at 72°C for 30s, 38 cycles, and finally extension at 72°C for 10min. aPCR system: PCR product (diluted 100 ti...
Embodiment 2
[0054] 1) Raw material
[0055] aPCR primer chain F (358EX-F) (5'→3'): GTCTTGAGTCCAACCCGG (18nt in length, SEQ ID NO: 1)
[0056] aPCR primer chain R (358EX-R) (5'→3'): ATGACCAAAATCCCTTAACG (5'-phosphorylated, 20nt in length, SEQ ID NO: 2)
[0057] splint(5'→3'):
[0058] 358B-S-20: CGTTAAGGGATTTTGGTCATGTCTTGAGTCCAACCCGGTA (40 nt, SEQ ID NO: 3)
[0059] 358B-S-25: ACTCACGTTAAGGGATTTTGGTCATGTCTTGAGTCCAACCCGGTAAGACA (50 nt, SEQ ID NO: 4)
[0060] 358B-S-30:
[0061] CGAAAACTCACGTTAAGGGATTTTGGTCATGTCTTGAGTCCAACCCGGTAAGACAC GAC (59nt, SEQ ID NO: 5)
[0062] Template source: Escherichia coli plasmid pUC18 (Thermo Fisher Scientific Biotechnology Co., Ltd.)
[0063] 2) Preparation of medium-length double-stranded DNA
[0064] 358nt double-stranded DNA was prepared by PCR. PCR system: [pUC 18]=2×104copies / μL, [358EX-F]=0.2 μM, [358EX-R]=0.2 μM, [Phanta Max Master Mix]=1×, total volume 20μL; pre-denaturation at 95°C 5min, denaturation at 95°C for 15s, annealing at 55°C for 15s,...
Embodiment 3
[0072] 1) Raw material
[0073] aPCR primer chain F (358EX-F) (5'→3'): GTCTTGAGTCCAACCCGG (18nt in length, SEQ ID NO: 1)
[0074] aPCR primer chain R (358EX-R) (5'→3'): ATGACCAAAATCCCTTAACG (5'-phosphorylated, 20nt in length, SEQ ID NO: 2)
[0075] splint(5'→3'):
[0076] 358B-S-20: CGTTAAGGGATTTTGGTCATGTCTTGAGTCCAACCCGGTA (40 nt, SEQ ID NO: 3)
[0077] Template source: Escherichia coli plasmid pUC18 (Thermo Fisher Scientific Biotechnology Co., Ltd.)
[0078] 2) Preparation of medium-length double strands
[0079] A 358bp double strand was prepared by PCR. PCR system: [pUC 18]=2×104copies / μL, [358EX-F]=0.2μM, [358EX-R]=0.2μM, [PhantaMax Master Mix]=1×, total volume 20μL; pre-denaturation at 95°C for 5min , Denaturation at 95°C for 15s, annealing at 55°C for 15s, extension at 72°C for 30s, 38 cycles, and finally extension at 72°C for 10min. Before the ligation reaction, the PCR product (monophosphorylated DNA double strand) was extracted to remove the PCR polymerase in th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com