Method for assisting in sorting lipidosome by utilizing single-stranded DNA nanostructure
A nanostructure and liposome technology, which is applied in liposome delivery, pharmaceutical formulations, medical preparations of non-active ingredients, etc., can solve the problem of uneven product, achieve high uptake rate, high cost, and good stability sexual effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] The sequence design of embodiment 1 single-stranded DNA nanostructure
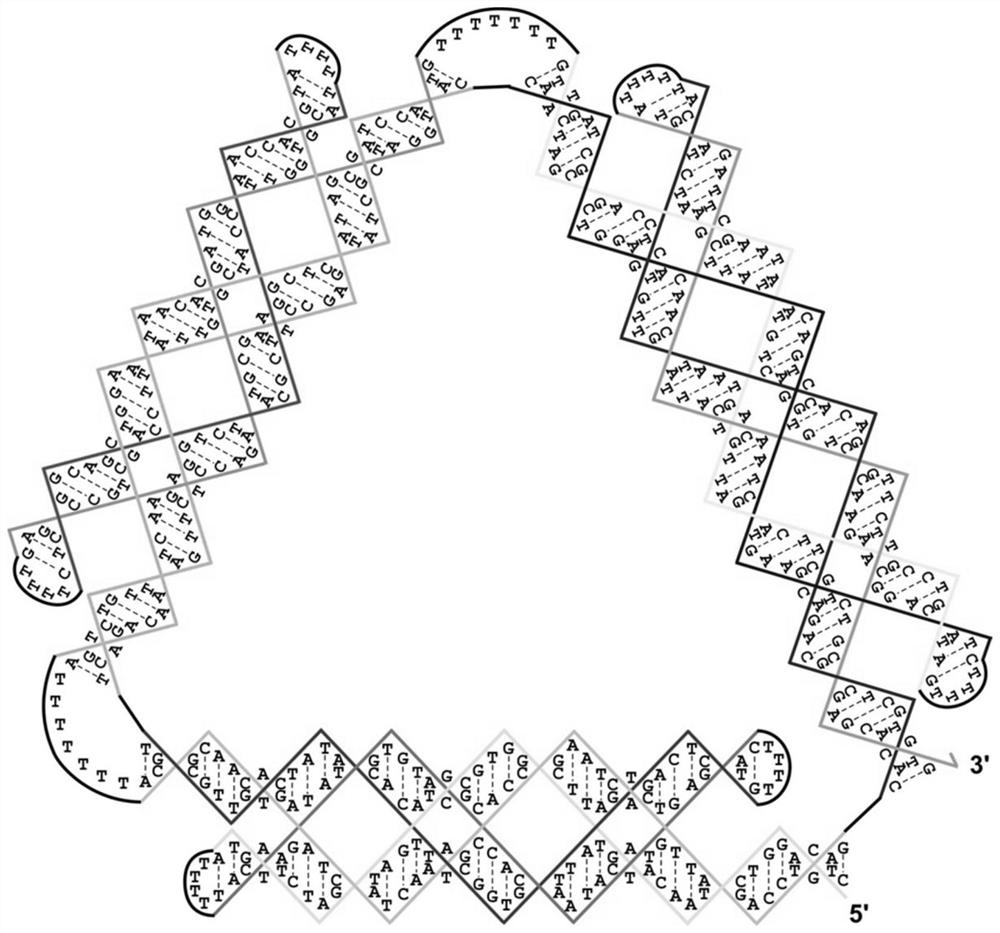
[0042] According to the design principle of the PX structure, taking a triangle as an example, two parallel double helices in each side are connected to another chain by crossing at a point close to each other. This structure can be formed by assembling a single-stranded DNA. The length of PX is designed to be between 10 and 20 nm on each side, so that its morphology can be clearly seen under the atomic force microscope. Then three PX molecules are covalently linked head-to-tail by 5'-3' to 5'-3' linkages to form triangles to ensure that the nanostructures to be generated can self-assemble from long ssDNA with a length of approximately 520 nt (e.g. figure 1 ), its sequence is shown in SEQ ID NO:1 sequence. Several TN (N=5 or 7) loops were placed at some tips to relax constraints in the edge-edge connection process.
Embodiment 2
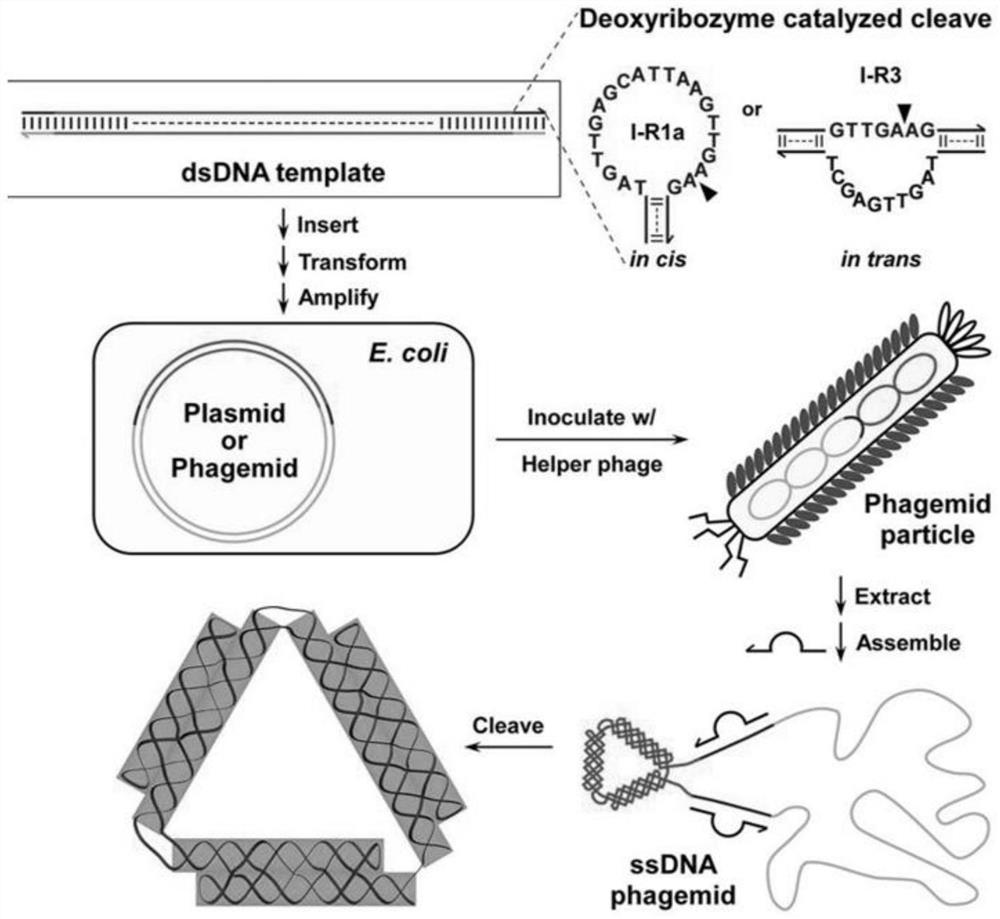
[0043] Example 2 Construction of phagemids comprising complete DNA nanostructure sequences and deoxyribozyme cleavage substrates
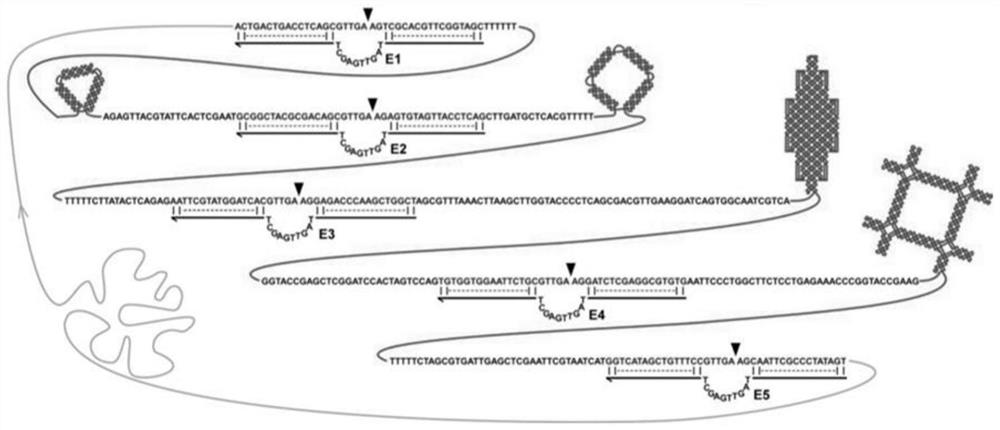
[0044] The complete sequence inserted into the phagemid vector consists of two parts: the deoxyribonase recognition site and the nanostructure sequence site. The nanostructure sequence is arranged in the middle, and the DNAzyme recognition sites are on both sides. Randomly select about 10 to 20 nucleotides as a spacer sequence and distribute it between the DNAzyme and the site of the DNA nanostructure to avoid steric hindrance between the two, thus ensuring the former in ssDNA amplification Even in the case of large structures in the vicinity, the daughter normally forms a hairpin structure and thus cuts normally. Taking the triangular structure as an example, a highly active self-cleaving DNAzyme (I-R1a) with a length of 45 nt (sequence: 5'-GATGTACAGCCATAGTTGAGCATTAAGTTGA / AGTGGCTGTACATC-3') was buried on both sides of the nanostructure site. Whe...
Embodiment 3
[0045] Example 3 Helper Phage Assisted Amplification of Single-Stranded DNA
[0046] A single p3024-triangle transformed colony was grown overnight at 37°C in 20 ml LB medium containing ampicillin (100 μg / mL). 2x YT medium (16.0g / L trypsin, 10.0g / L yeast extract, 5.0g / L NaCl, 5mM MgCl 2 ) were inoculated with 3 mL of overnight culture, and shaken at a speed of 250 r / min. When the OD600 of the culture reached about 0.4-0.5, VCSM13 helper phage was added thereto to an MOI of 20. After 30 min, kanamycin (final concentration 50 μg / mL) was added to the culture to select infected cells. After about 4.5 hours, the culture was collected and centrifuged at 4000 rcf for 15 minutes at 4°C to remove bacteria. The supernatant was transferred to a clean bottle and dissolved with PEG 8000 (40 g / L) and NaCl (30 g / L). The mixture was incubated on ice for 30 minutes and then centrifuged at 5000 rcf for 30 minutes at 4°C. The supernatant was discarded and the phagemid pellet was resuspended ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| The average diameter | aaaaa | aaaaa |
| The average diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com