Application of KMT2D in preparation of antitumor drugs
An anti-tumor drug, KMT2D-LCD1 technology, is applied in the application field of KMT2D in the preparation of anti-tumor drugs, and can solve the problems of unreported functions and unclear tumor functions.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] 1. Structural analysis of KMT2D protein and determination of LCD domain
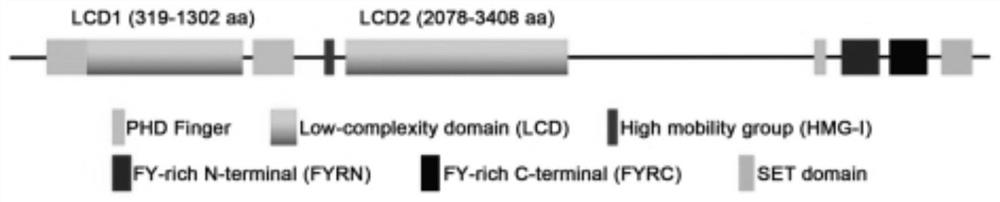
[0044]Referring to the relevant information in the NCBI database, the amino acid sequence of Histone-lysine N-methyltransferase 2D (KMT2D) (GenBank: NP_003473.3) was obtained, and the KMT2D protein structure mainly included two clusters of PHDFinger regions (3) and one at the N-terminus PHD Finger, N-terminal contains FYRN and FYRC domains, and also contains a core methyltransferase structure SET region, its structure is as follows figure 1 As shown, the full-length KMT2D protein contains 5537 amino acids, constituting the core structure of the KMT2D methylation enzymatic complex.
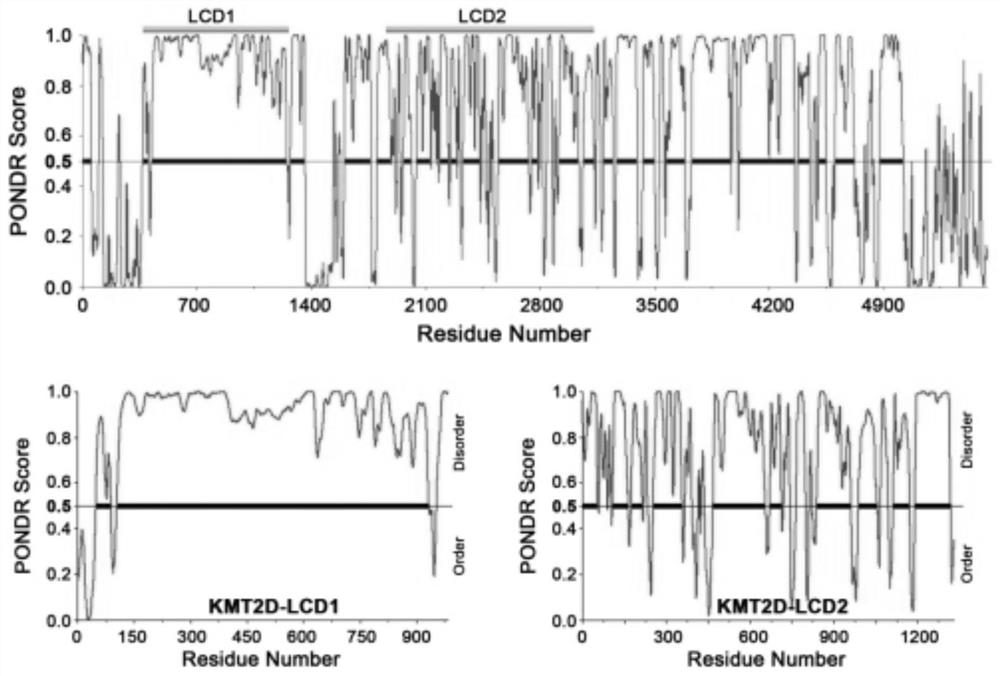
[0045] The disordered structure of the KMT2D protein was analyzed by the Predictor of Natural Disordered Regions software, and it was found that there were two highly disordered regions in the KMT2D protein, such as figure 2 Shown, namely LCD1 and LCD2. Further analysis found that the LCD1 structure is located at 31...
Embodiment 2
[0049] 1. The knockout strategy of LCD1 domain and its verification
[0050] Aiming at the first disordered protein structure in KMT2D protein, that is, LCD1, which is located in the 8th exon to the 11th exon in its corresponding gene, two sgRNAs (sgRNA A1 and sgRNA A2), and two sgRNAs (sgRNA B1 and sgRNA B2) are designed downstream. The sgRNA sequence is as follows:
[0051] sgRNA A1: ccggggacaatagggcagaatca (SEQ ID NO. 1)
[0052] sgRNA A2: ccgggagtgggcaaaacaggcat (SEQ ID NO. 2)
[0053] sgRNA B1: ccggtgaggggctatctagctgc (SEQ ID NO. 3)
[0054] sgRNA B2: ccgcaggactgtacctctgacag (SEQ ID NO. 4).
[0055] Such as Figure 4 As shown, the LCD1 region was knocked out in 293T and PANC1 cells using CRSPR Cas9 gene editing technology.
[0056] 1.1 Synthesis of sgRNA
[0057] The sequence of the designed sgRNA primer was synthesized (Suzhou Jinweizhi Biotechnology Co., Ltd.), and the vector epiCRISPR was cut through the specific enzyme cutting site BspQI. Then use T4 ligase to...
Embodiment 3
[0112] 1. Effect of LCD domain on tumor cell proliferation
[0113] Using the constructed LCD1 or LCD2 knockout stable cell lines, CCK8 was used to detect the proliferation ability of various cells. The specific implementation steps are as follows:
[0114] (1) Inoculate the suspension of stably transfected cells (HEK293T or PANC1) in a 96-well plate, pre-culture for 24 hours, and set up a blank group and a control group at the same time;
[0115] (2) Add 10 μL of CCK-8 Solution to each well, add along the cell plate wall, mix gently, do not produce air bubbles, and incubate in the incubator for 2 h;
[0116] (3) Measure the absorbance at 450 nm with a microplate reader.
[0117] During the experiment, the relative activity of the cells was detected on the 0th day, the 1st day, the 3rd day and the 5th day, and the results were as follows: Figure 12 and Figure 13 shown. Results analysis, in HEK293T ( Figure 12 ), the cell proliferation of the LCD knockout group (includ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com