Alpaca-derived nanobody binding to SARS-CoV-2 RBD
A sars-cov-2rbd, antibody technology, applied in the direction of antibodies, antiviral agents, hybrid immunoglobulins, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0107] Example 1 Using SARS-CoV-2 RBD to Immunize Alpacas and Screen Nanobodies
[0108]1) The purified SARS-CoV-2 RBD (QKV42562.1, aa 321-591) expressed in HEK293F cells (ATCC, CBP60437) was mixed with Freund's adjuvant, and the alpaca was subcutaneously injected with 500 μg / time for three times, Two 6-month-old female alpacas were vaccinated at intervals of 2 weeks.
[0109] 2) Two weeks after the third immunization, blood was collected from a vein and white blood cells in the blood were separated. Total RNA was extracted using the RNA extraction kit from Omegabiotek, and genomic DNA was removed using DNase. Using TAKARA's PrimeScript TM II 1st Strand cDNA Synthesis Kit performs reverse transcription on RNA and reverse transcribes RNA into cDNA.
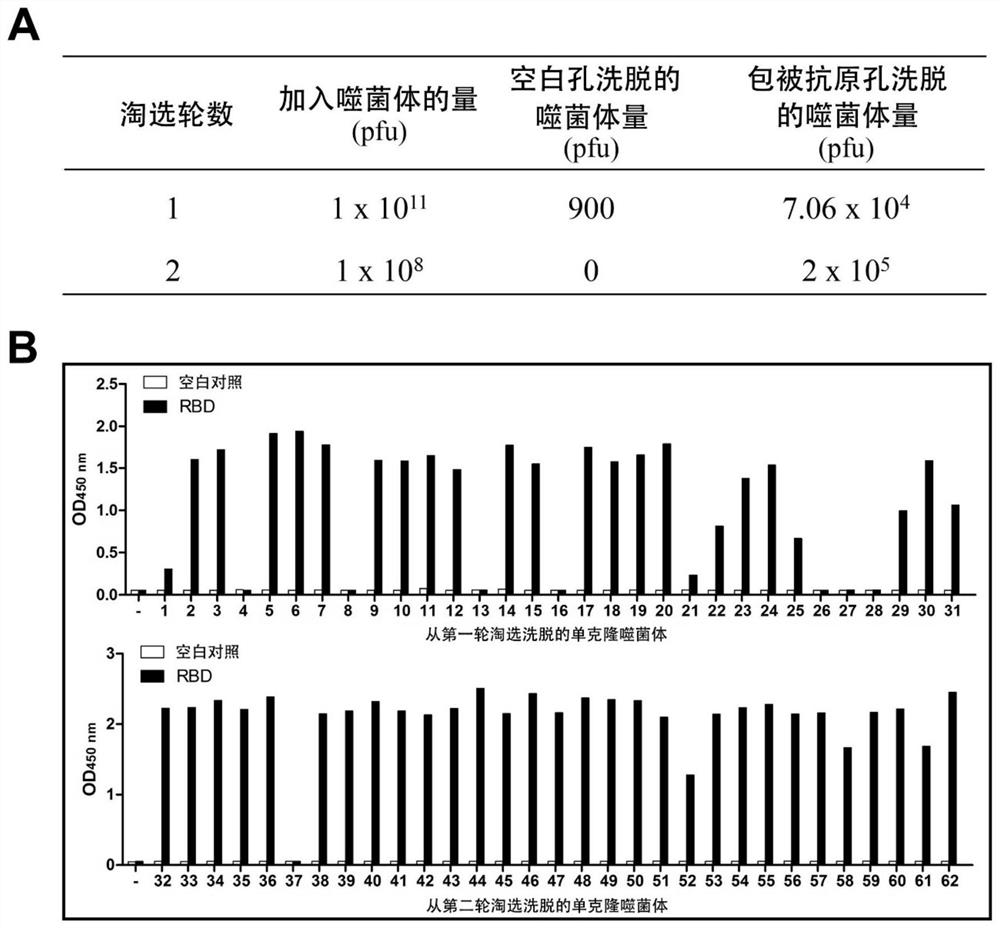
[0110] 3) Preparation of nanobody phagemid library: use the alpaca VHH-specific primers we designed to amplify the coding gene fragment of VHH using the above cDNA as a template, and clone the amplified VHH sequence into a phag...
Embodiment 2
[0121] Example 2 Expression and purification of nanobodies and their Fc fusion proteins
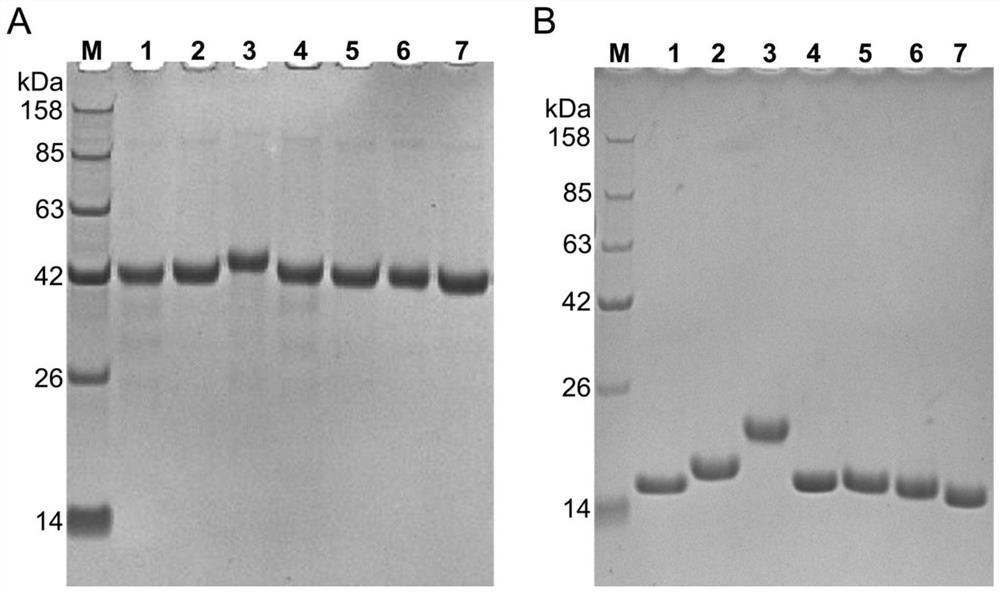
[0122] 1) Design primers, fuse the N-terminus of the gene sequence of the Nanobody to IFNα protein signal peptide to guide secreted expression, fuse the C-terminus of the gene sequence of the Nanobody to human IgG1 Fc, and introduce a TEV restriction enzyme between them site, and then cloned into the mammalian expression vector pTT5. The constructed vector was transiently transfected into mammalian HEK293F cells with PEI, and the supernatant was collected after 3 days of culture. The fusion protein in the supernatant was purified by Protein A column, and SDS-PAGE electrophoresis was performed. The results are as follows: figure 2 As shown in A, from the supernatant, we obtained highly pure Nanobody Fc fusion protein.
[0123] 2) Digest the fusion protein with TEV, then flow the digested products through Protein G column and nickel column respectively, so as to remove the undigested prot...
Embodiment 3
[0124] Example 3 Characterization of the Nanobodies
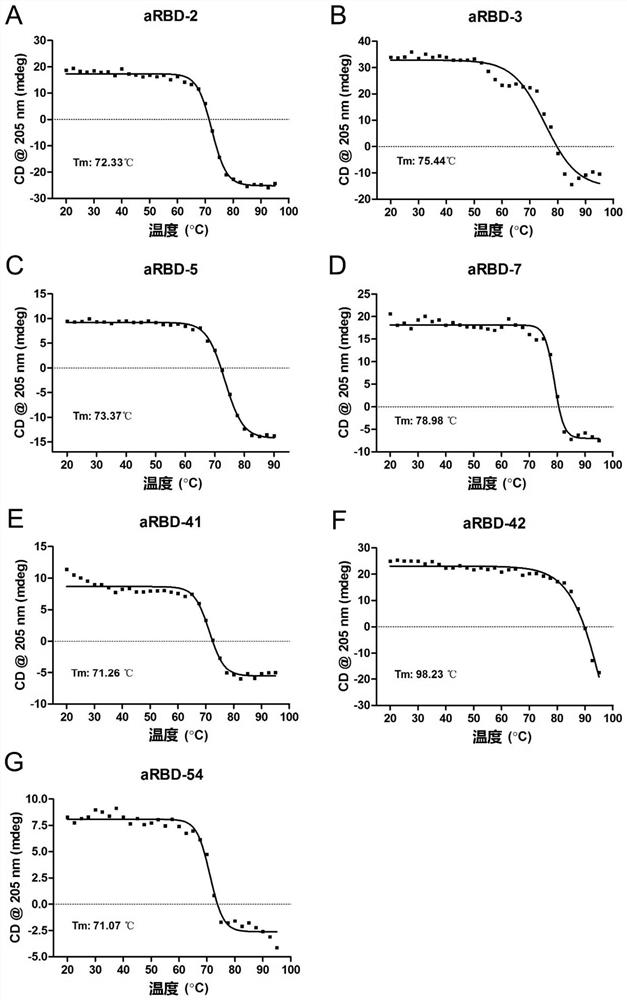
[0125] 1) Use circular dichroism (CD) to characterize the stability of nanobodies: replace the nanobody solutions of the examples with PBS and dilute to OD 280nm It is about 0.6, and then detected by a circular dichroism spectrometer, the detection wavelength range is 280nm-180nm, and the temperature is from 20-95°C. Each assay was repeated twice. Prism software was used to process the data, and the variation of the spectral value at 205 nm with temperature was selected, and the Tm value was further fitted. The result is as image 3 As shown, the Tm values of aRBD-2-Fc, aRBD-3-Fc, aRBD-5-Fc, aRBD-7-Fc, aRBD-41-Fc, aRBD-42-Fc and aRBD-54-Fc were 72.33 , 75.44, 73.37, 78.98, 71.26, 98.23 and 71.07°C.
[0126] 2) Preliminary characterization of the binding of the Nanobody Fc fusion protein to the extracellular segment of the SARS-CoV-2 spike protein (S1+S2) by non-competitive ELISA: the SARS-CoV-2 SARS-CoV-2 spike protei...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com