Method for detecting inhibition for bacteria by antibiotics through single-cell counting
A technology of bacteria inhibition and antibiotics, applied in the field of biomedicine, can solve problems such as complicated operation, expensive equipment, and difficult judgment of practical prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Objective: To search for the sensitive indicators of bacterial changes during bacterial culture in broth, to determine the theoretical basis for rapid drug susceptibility testing, and to establish a detection method for bacterial drug susceptibility.
[0041] Materials and methods:
[0042] 1. Strain preparation:
[0043]Three standard bacterial strains (ATOC 25922 Escherichia coli, ATOC 25923 Staphylococcus aureus, ATOC 27853 Pseudomonas aeruginosa) were transferred for use, and incubated at 37° C. for 18 hours.
[0044] 2. Broth preparation:
[0045] Prepare bacterial strain and grind on the bottle wall of AST (antibiotic susceptibility test) broth, mix well, cover the bottle cap and measure turbidity with a turbidimeter (BD company Phoeni xSpec Nephel omter), the turbidity is 0.5 McFarland units, spare. Inoculate the bacteria to be tested according to the inoculum concentration of the CLSI (American Clinical Laboratory Standards Organization) broth dilution method...
Embodiment 2
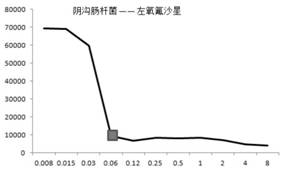
[0060] Ampicillin inhibition detection method for ATOC 25922 Escherichia coli (using resistance counting method):
[0061] Materials and methods:
[0062] 1. Strain preparation:
[0063] The standard strain ATOC 25922 Escherichia coli was transferred for use and incubated at 37°C for 18 hours.
[0064] 2. Broth preparation:
[0065] 10 of the drug susceptibility test tubes (or cups) contain the required antibiotics at the double dilution concentration (different drug concentrations refer to the US CLSI standard); the eleventh tube does not contain antibiotics, as a positive control (PC); there is no bacterial suspension The twelfth tube was used as a negative control (NC).
[0066] The prepared bacterial strains are ground on the bottle wall of MH (antibiotic susceptibility) broth, mixed evenly, and the bottle cap is closed and the turbidity is measured with a turbidimeter (BD company Phoeni xSpec Nephel omter), the turbidity is 0.5 McFarland units, and the .
[0067] Ino...
Embodiment 3
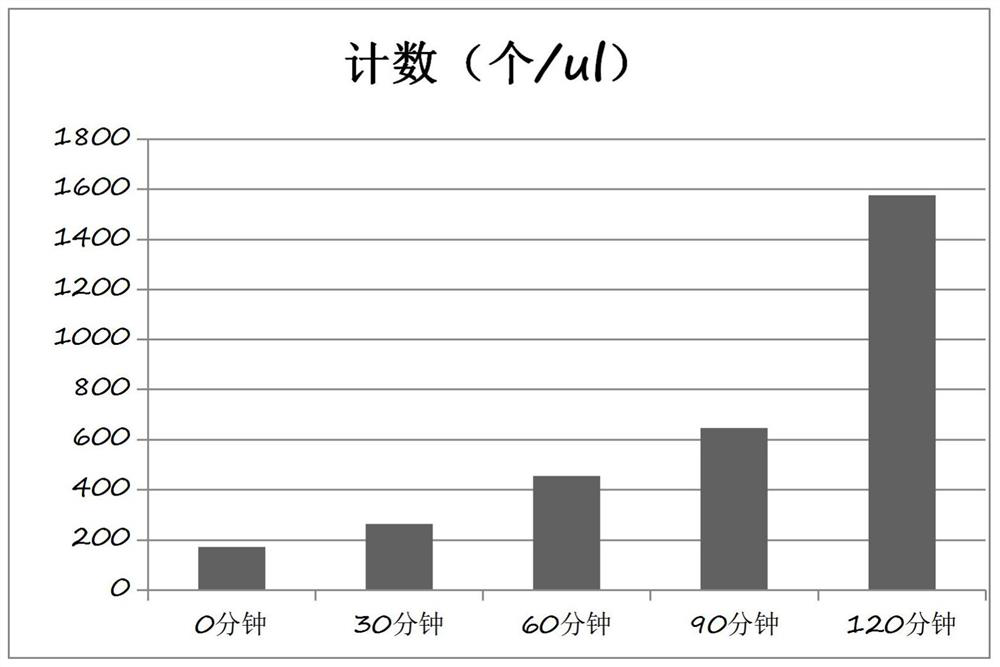
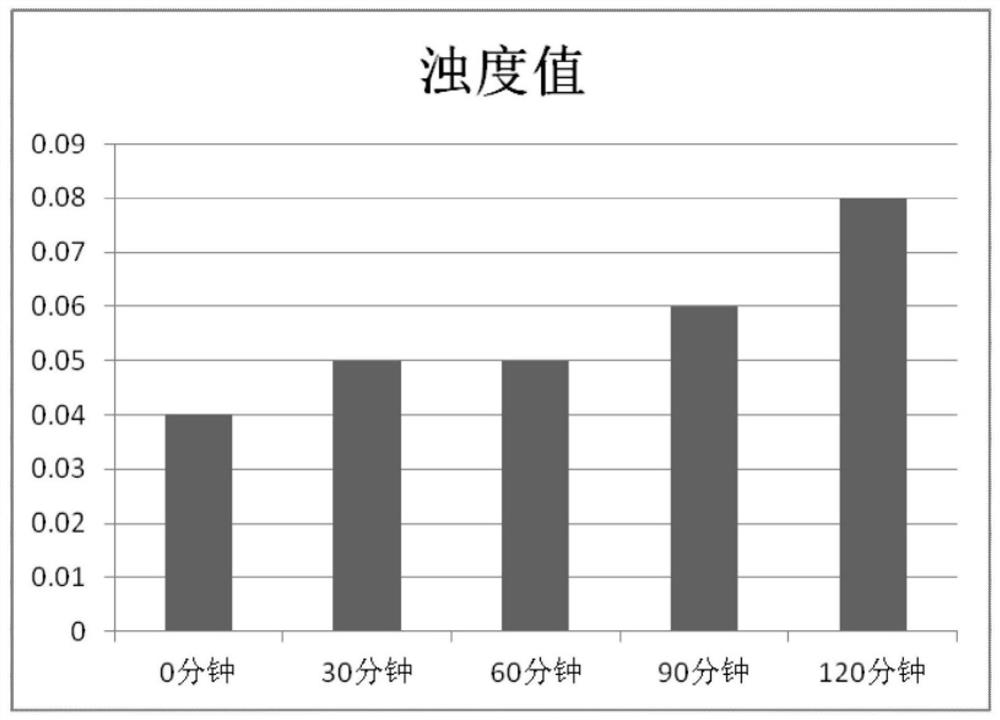
[0084] 1. Growth experiment
[0085] 1.1 Purpose of the experiment
[0086] According to the requirements of CLSI standard, inoculate the six common clinical strains (ATOC29212, ATOC29213, ATOC27853, ATOC25922, Klebsiella pneumoniae ATOC700603 and Acinetobacter baumannii) and record their growth trend to determine whether there is a possibility of their growth within 2 hours sex.
[0087] 1.2 Experimental method
[0088] 1.2.1 Negative control: test the uninoculated medium, use a resistance bacteria counter to test, and record the number of particles;
[0089] 1.2.2 Strain preparation: According to the requirements of CLSI standards, pick 0.5 McFarland fresh strains within 24 hours, take 100ul and add it to 10ml of culture medium;
[0090] 1.2.3 Record the results at 0h / 0.5h / 1h / 1.5h respectively, remove the negative background and analyze the results.
[0091] 2.3 Experimental results see Figure 4 , It can be seen from the results of the growth experiment that the bacter...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com