qRT-PCR reference genes suitable for Rehmannia chingii Li and application of qRT-PCR reference genes
A technology of Tianmu Dihuang and internal reference gene, which is applied to qRT-PCR internal reference gene and its application field, can solve the problems that cannot be used as Tianmu Huang internal reference gene, internal reference gene has no universality, lack of Tianmu Huang internal reference gene, etc., achieves high sensitivity, Good specificity and good stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Embodiment 1: Extraction of plant total RNA and synthesis of cDNA
[0032] Total RNA was extracted from all test samples according to the instructions of the RNA extraction kit (TaKaRa, Dalian), and the contamination of genomic DNA was removed by DNase treatment. The concentration and quality of the total RNA were measured by an ultra-micro nucleic acid protein analyzer, and the integrity of the total RNA was detected by 1% agarose gel electrophoresis.
[0033] The OD260 / OD280 and OD260 / OD230 of each test sample RNA met the requirements, and the agarose gel electrophoresis test showed that the 28S and 18S bands were clear without degradation, which met the requirements.
[0034] cDNA was synthesized by reverse transcription using reverse transcription kit type 6210A (TaKaRa, Dalian). About 1 μg of total RNA, 1 μL of oligo dT primer (concentration: 50 μmol L -1 ), 1 μL dNTP, 0.5 μL RNase inhibitor (40U·μL -1 ), 1 μL PrimeScript II reverse transcriptase (200U·μL -1 ),...
Embodiment 2
[0036] Example 2: Selection of candidate internal reference genes and primer design
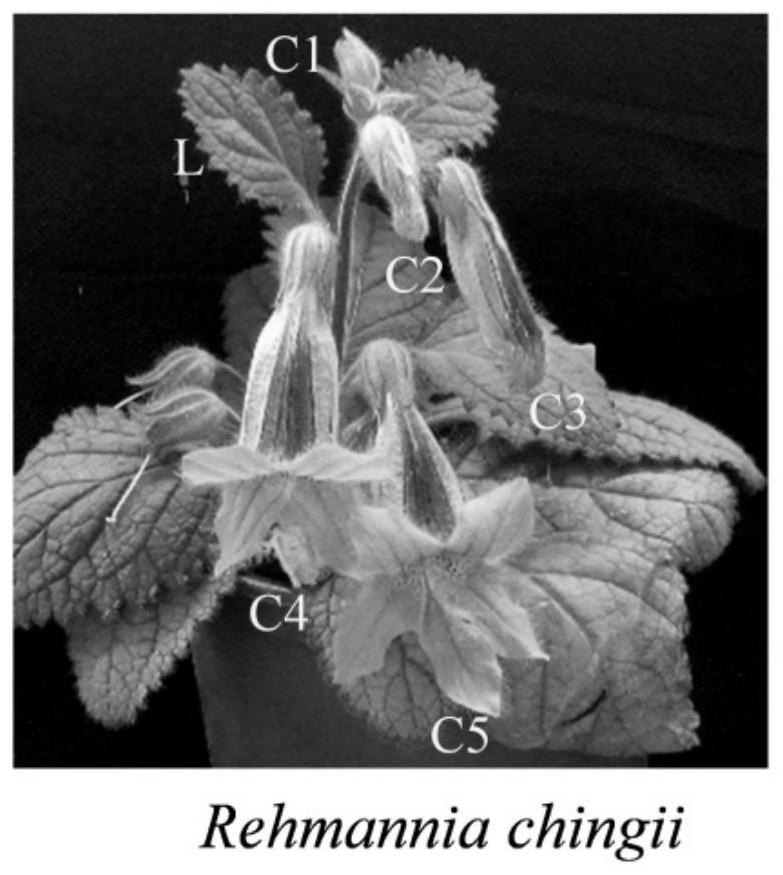
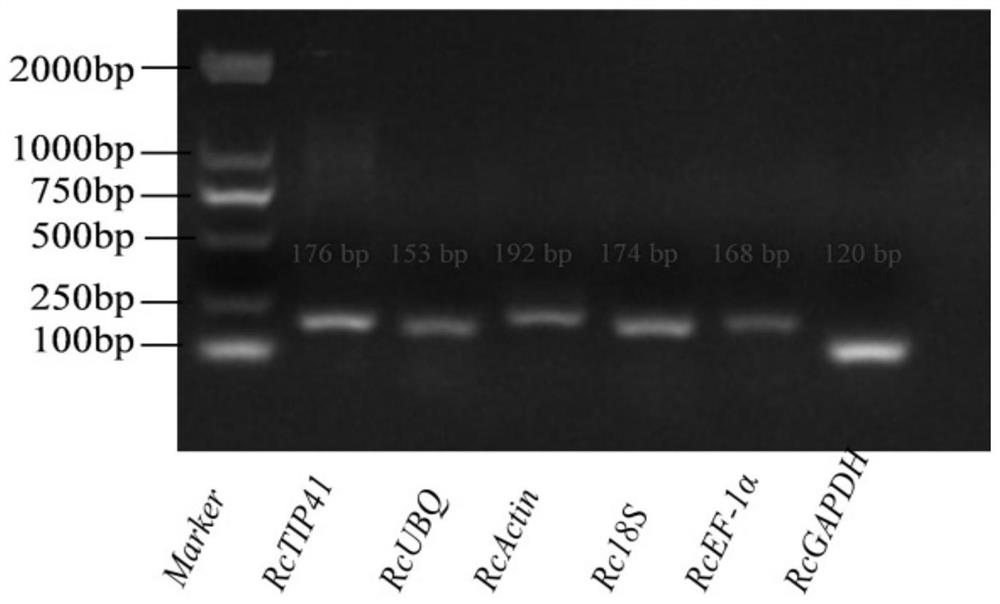
[0037]According to the transcriptome sequencing data of Tianmudi Huang flowers and leaves, six different candidate internal reference genes were screened, namely RcTIP41, RcUBQ, RcActin, Rc18S, RcGAPDH and RcEF-1α, in all tested samples (flowers at different developmental stages: The FPKM values of these six genes were relatively stable in the small bud stage (C1), budding stage (C2), big bud stage (C3), early flowering stage (C4), full flowering stage (C5) and leaf (L) ( Table 1). Among them, FPKM refers to the fragments of transcription per thousand bases per million mapped reads. This value can reflect the expression level of the gene. The larger the FPKM value, the higher the expression level of the gene. NCBI online analysis software ORFFinder was used to predict the open reading frame of the cDNA sequence of each gene. Then, according to the sequence of the gene, quantitative specif...
Embodiment 3
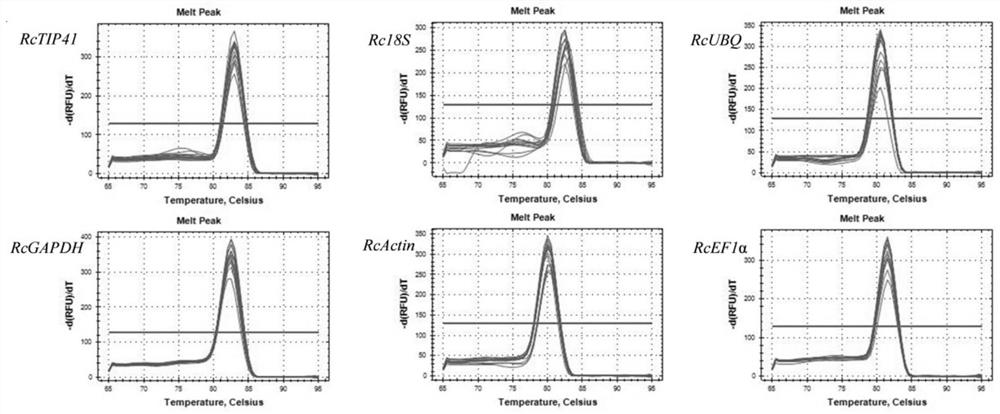
[0043] Example 3: Stability analysis of candidate internal reference genes
[0044] Using the 10-fold diluted cDNA as a template, according to the Premix Ex TaqTM II (TliRNaseH Plus) (TaKaRa, Dalian) kit instructions prepared qRT-PCR reaction system, each reaction was repeated 3 times, and carried out on the 96-well plate of BIO-RAD IQ5 quantitative instrument (Bio-Rad, Shanghai) Real-time fluorescence quantitative PCR.
[0045] The reaction system is: 12.5 μL of Premix Ex enzyme, 1 μL forward primer (10 μmol L-1), 1 μL reverse primer (10 μmol L-1), 1.0 μL cDNA template, plus 9.5 μL deionized water, the total volume is 25 μL.
[0046] The qRT-PCR reaction program was: 95°C for 30s; 95°C for 5s, 60°C for 30s, 40 cycles.
[0047] The expression level of candidate internal reference genes can be initially evaluated by the original Cq value obtained by qRT-PCR. The larger the Cq value of the gene, the lower the expression level of the gene, and vice versa. The stability of c...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com