Systems and methods for genetic manipulation of akkermansia species
A kind of Akkermansia gene technology, applied in Akkermansia bacteria, genetic modification Akkermansia bacteria, treatment of diseases, can solve the problem of Akkermansia difficult molecular manipulation and so on
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
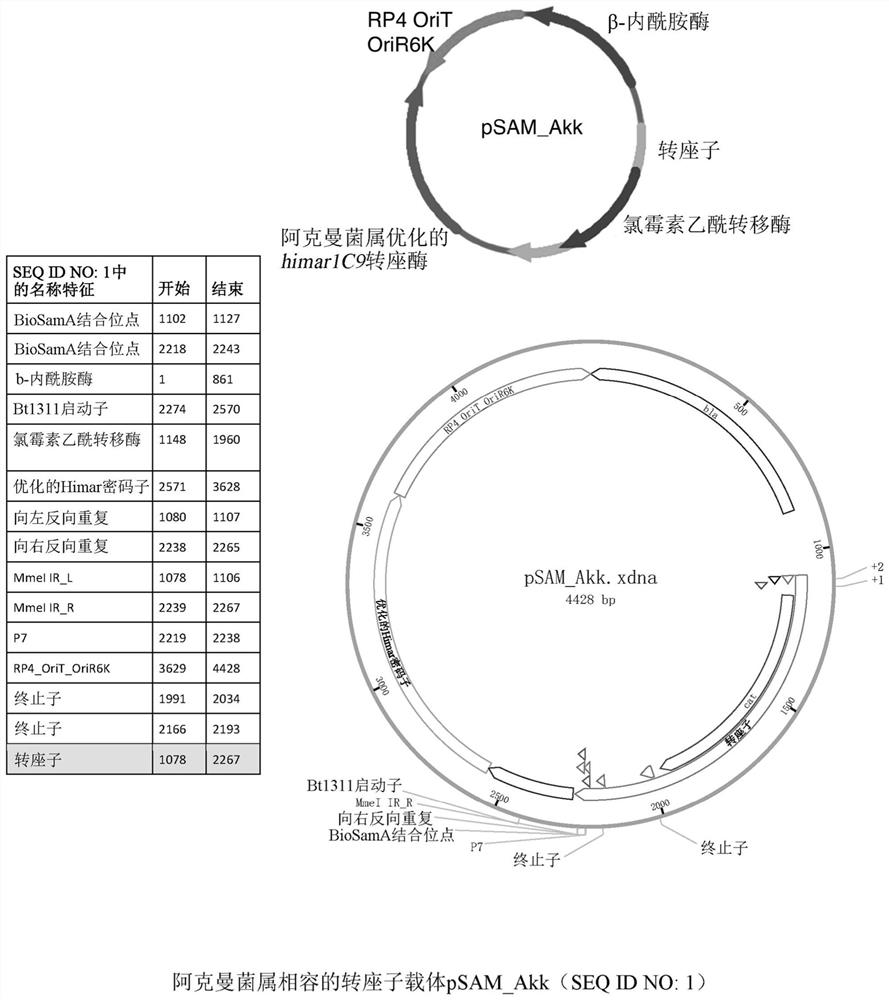
[0078] Example 1: Modified Tn mutagenesis vector for use in Akkermansia: pSAM_Akk
[0079] To enable genetic screening for genes with altered phenotypes and altered bacteria, tools for mutating bacteria are needed. A modified version of the previously described vector was generated for use in Akkermansia. Original vector pSAM_Bt 1 Designed for use with Bacteroides thetaiotaomicron. The vector encodes both the modified mariner transposon himar1C9 with the erythromycin resistance gene and the transposase required to catalyze transposition. The plasmid uses a lamba pir-dependent origin of replication and cannot replicate in strains lacking the pir gene such as Akkermansia.
[0080] For compatible use of pSAM_Bt in Akkermansia, the original erythromycin resistance marker (ermG) on the transposon was replaced with a chloramphenicol resistance cassette (cat). Initial attempts to use erythromycin as a selectable marker in Akkermansia were unsuccessful, and growth with erythromyci...
example 2
[0081] Example 2: Methods for Mutagenesis and Transposon (Tn) Library Construction
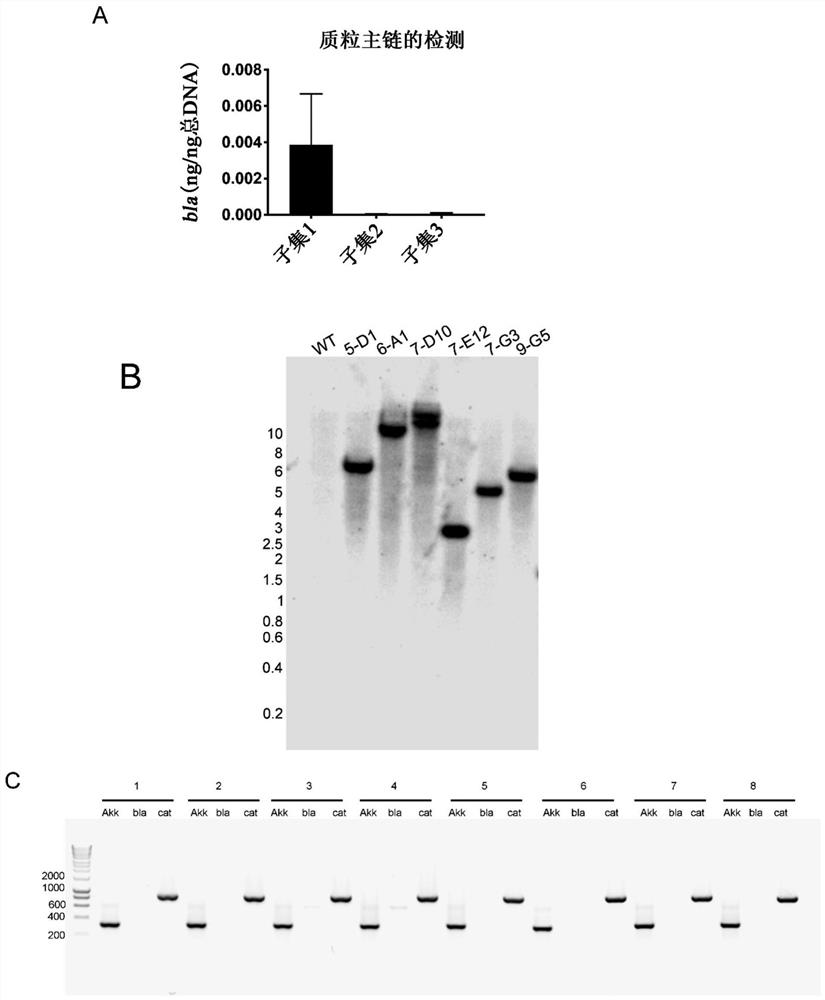
[0082] The transposon vector (SEQ ID NO: 1 ) was introduced into Akkermansia by conjugation to an E. coli donor strain. Akkermansia starter cultures were subcultured at a ratio of 1:5 in 30ml of synthetic medium 2 Medium and grow to OD600=0.6-1.0. Cells were then harvested by centrifugation at 10 000 xg in 1.5 ml tubes for 5 minutes at 4°C. At the same time, the Escherichia coli S17 pSAM_Akk culture was aerobically cultured in LB+100ug / ml ampicillin, 35ug / ml chloramphenicol at 37°C and 200rpm until the optical density (OD) OD600=0.4-0.7. To avoid shearing of conjugated pili, E. coli were centrifuged at 2000 xg for 3 minutes and washed once with sterile PBS. The E. coli and Akkermansia pellets were combined in synthetic medium to a total volume of 0.5 ml, and the suspension was used to make 100 μl puddles on pre-reduced synthetic medium plates. Plates were incubated aerobically at 37°C for ...
example 3
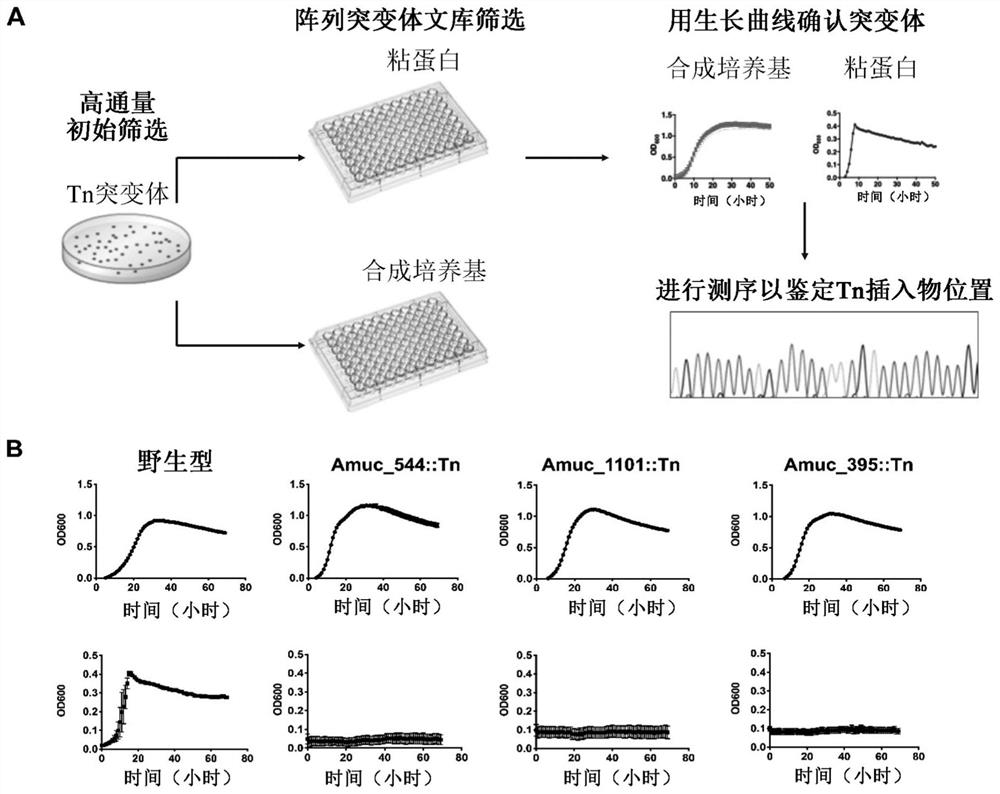
[0085] Example 3: Transposon mutant screening - transposon mutants were screened for genes required for mucin utilization
[0086] To screen for genes required for mucin utilization, array Tn mutants were used to inoculate mucin-containing media 3 Or replicate 96-well plates with synthetic media. Plates were incubated anaerobically at 37°C for 3 days. After growth, measure OD600 using a microplate reader. Mutants that grew in synthetic media but not in mucin were selected for additional characterization. To confirm the initial screen, mutants of interest were tested for mucin growth defects by performing a growth curve assay in a microplate reader, with measurements taken every 60 min for 72 h ( image 3 ). Random PCR was used to map transposon insertion sites and identify genes required for growth on mucins. The screen led to the identification of genes specifically required for growth on mucins but not on monosaccharides (Table 1).
[0087] Table 1. Genes identified as...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com