Method for identifying traditional Chinese medicinal material rhizome pinelliae and easily-confused products thereof
A technology of Chinese medicinal material Pinellia, which is applied in the field of identification of Chinese medicinal material Pinellia and its easily mixed products, can solve problems such as difficult to effectively and accurately identify Pinellia and its easily mixed products, and achieve shortened analysis time and cost, accurate Identify and reduce the effects of sequencing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
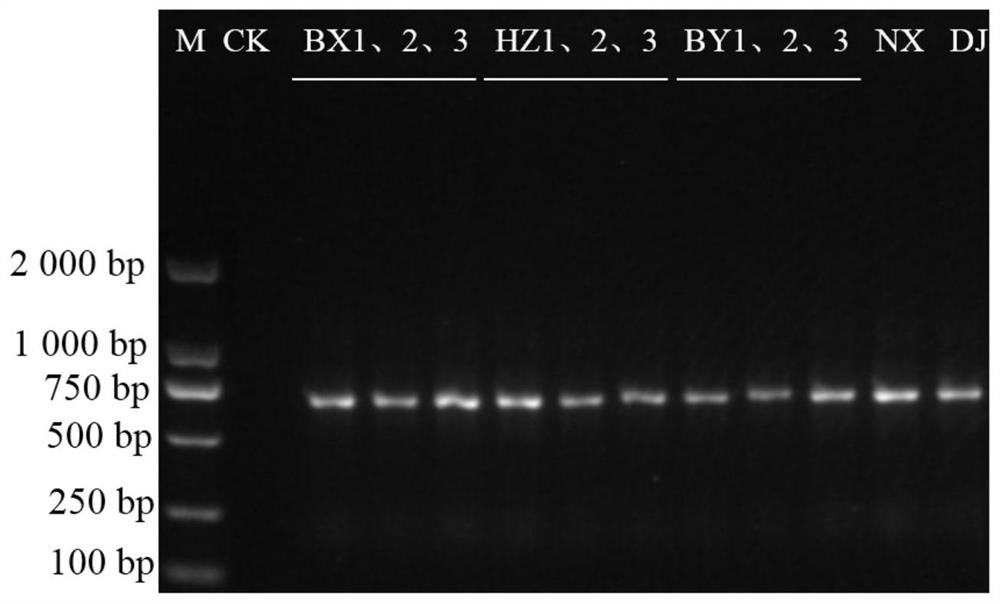
[0093] Embodiment 1: the primer pair of pinellia and easy-to-mix product is first PCR amplification
[0094] 1. Sample source
[0095] The authentic pinellia used in this example (fresh or dry products can be used, fresh products need to be dried and crushed at 45°C), common easily mixed products such as tiger palm, whip eaves plowshare tip, other easily mixed products Aracea, unicorn lotus From Hebei, Hunan, Anhui and Guangzhou.
[0096] 2. DNA extraction
[0097] DNA extraction was performed on the sample, and the extraction method was as follows:
[0098] 1) Pulverization: sterilize the surface of the sample with 75% ethanol, cut the sample into small pieces, and grind it into powder with a high-throughput tissue grinder;
[0099] 2) Removing starch and polysaccharides: Take 100-150 mg of the pulverized sample, mix with 1 mL of nuclear separation solution, centrifuge at a low speed of 3 500 r / min for 5 minutes, repeat the operation of adding liquid and centrifugation for...
Embodiment 2
[0116] Example 2: Amplification of Multiplex Site-Specific PCR Primer Pairs
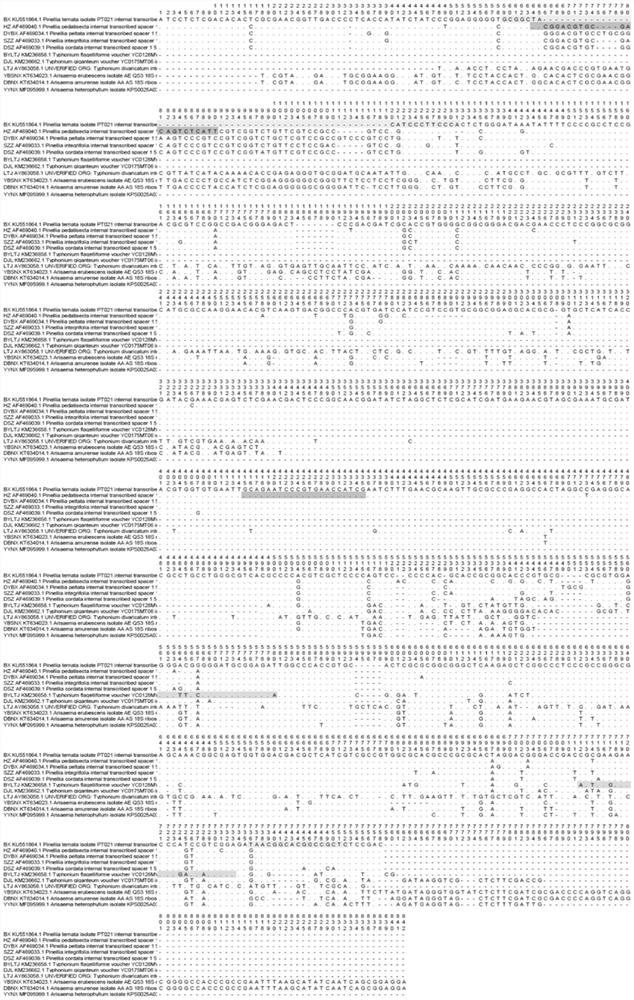
[0117] 1. Design multiple site-specific PCR primer pairs
[0118] Download the ITS sequences of Pinellia genus Pinellia, Tiger Palm, Dripdrop, Stone Spider and Pinellia scutellaria from Genbank (accession numbers: KU551864.1, AF469040.1, AF469039.1, AF469033.1, AF469034.1), The ITS sequences of the plants of the genus Aracea, the eaves of the eaves, the unicorn, and the unicorn (accession numbers: KM236658.1, AY863058.1, KM236662.1), the ITS sequences of the plants of the genus Aracea, Aracea heterophyllum, and Aracea dongbei ITS sequence (accession numbers: KT634023.1, MF095999.1, KT634014.1).
[0119] The multiple sequence alignment analysis of the ITS sequences of the above plants was carried out using Mega X software. The ITS sequence of pinellia is A at 77bp, the others are T, the ITS sequence at 662bp is T, and the others are G; the ITS sequence of Tiger palm is T at 84bp, the others are G, C...
experiment example 1
[0141] Experimental Example 1: Process Screening
[0142] 1. Download the ITS sequences of Pinellia, Pinellia and Pinellia scutellaria from Genbank (accession numbers: KU551864.1, AF469040.1, AF469039.1, AF469033.1, AF469034.1) The ITS sequences of the plants Whipcornia, Plowshare and Unicorn (accession numbers: KM236658.1, AY863058.1, KM236662.1), the ITS sequences of Araceae plants Aracea, Heterophyllum and Northeast Aracea (accession numbers No.: KT634023.1, MF095999.1, KT634014.1).
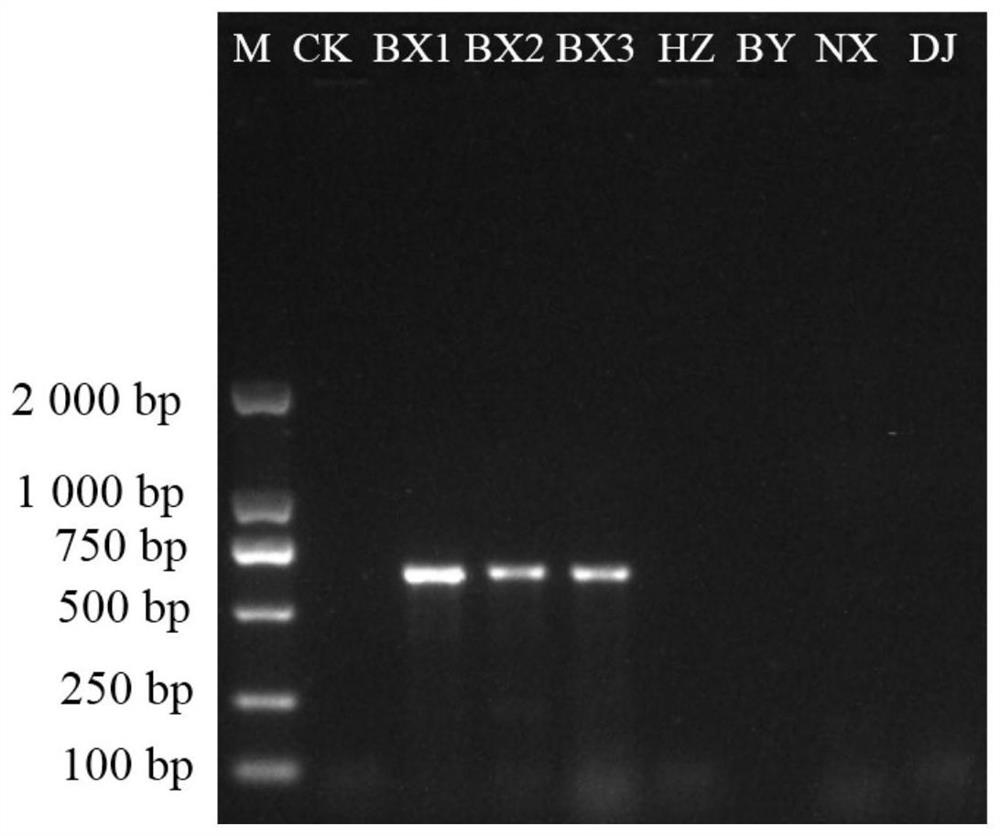
[0143] The multiple sequence alignment analysis of the ITS sequences of the above plants was carried out using Mega X software. The designed primers are listed in Table 2.
[0144] Table 2 Pairs of specific PCR primers for pinellia chinensis, tiger palm and whip eaves
[0145]
[0146]
[0147] Based on the fact that "the specific primers designed for a variety only have specific bands for this variety, and other easily mixed products have no amplified bands", after PCR amplification ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com