Yeast genome editing method based on CRISPR/Cas9
A genome editing and genome technology, applied in microorganism-based methods, other methods of inserting foreign genetic materials, genetic engineering, etc., can solve the problems of low PCR product concentration, unfavorable operation, etc., to ensure editing efficiency, shorten experimental time and Cost, efficient and convenient editing effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Embodiment 1, the construction of expressing Cas9 bacterial strain YD005
[0043] A major player in glucose repression is a zinc finger protein, MIG1, that binds to the promoters of many genes and represses their transcription by repressing genes encoding transcriptional activators. The inhibitory effect of MIG1 can be direct or indirect. Galactose metabolism is controlled by an enzyme encoded by the GAL gene, however, in the presence of glucose, the GAL gene is repressed by a repressor gene. Most of the GAL genes are directly repressed, while the GAL4 gene is indirectly repressed through MIG1 (Jakub et al. Combinatorial control of gene expression by the three yeast repressors Mig1, Mig2 and Mig3[J]. Bmc Genomics, 2008.). In order to improve the activity of the GAL-type promoter, we knocked out MIG1 and at the same time realized the expression of Cas9 protein, which provided the basis for subsequent gene editing.
[0044] 1. Construction and transformation of Cas9 pro...
Embodiment 2
[0053] Example 2 Preparation of GRE3, PHO13 and ASC1 gRNA expression cassettes
[0054] 1. Construction of gRNA expression cassette containing target site GRE3
[0055] Utilize chopchop website, design target sequence according to target site GRE3 gene (Saccharomyces Genome DatabaseYHR104W), described target sequence is as follows:
[0056] GRE3 target sequence: ACCAATCATAGATACGTACC (SEQ ID NO. 10).
[0057] Using Addgene's commercialized plasmid p426-SNR52p-gRNA.CAN1.Y-SUP4t (abbreviated as p426) as a template, primers P11 / P12 were used to amplify the upstream homology arm fragment of the gRNA expression cassette of the target site GRE3 ( SEQ ID NO.1); Using primers P13 / P14, PCR amplification obtained the downstream homology arm fragment (SEQ ID NO.5) of the gRNA expression cassette of the target site GRE3. The DNA splicing method is as described in Example 1. The upstream homology arm fragment, SNR52p promoter fragment (SEQ ID NO.2), target sequence (SEQ ID NO.10), gRNA Sc...
Embodiment 3
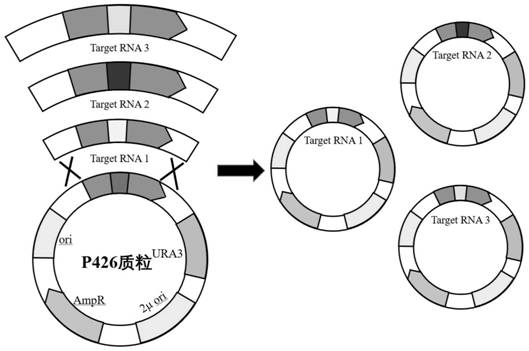
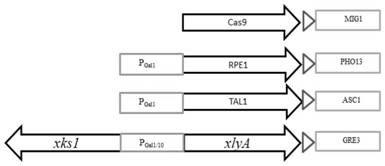
[0069]Embodiment 3, transformation of genes related to xylose metabolic pathway
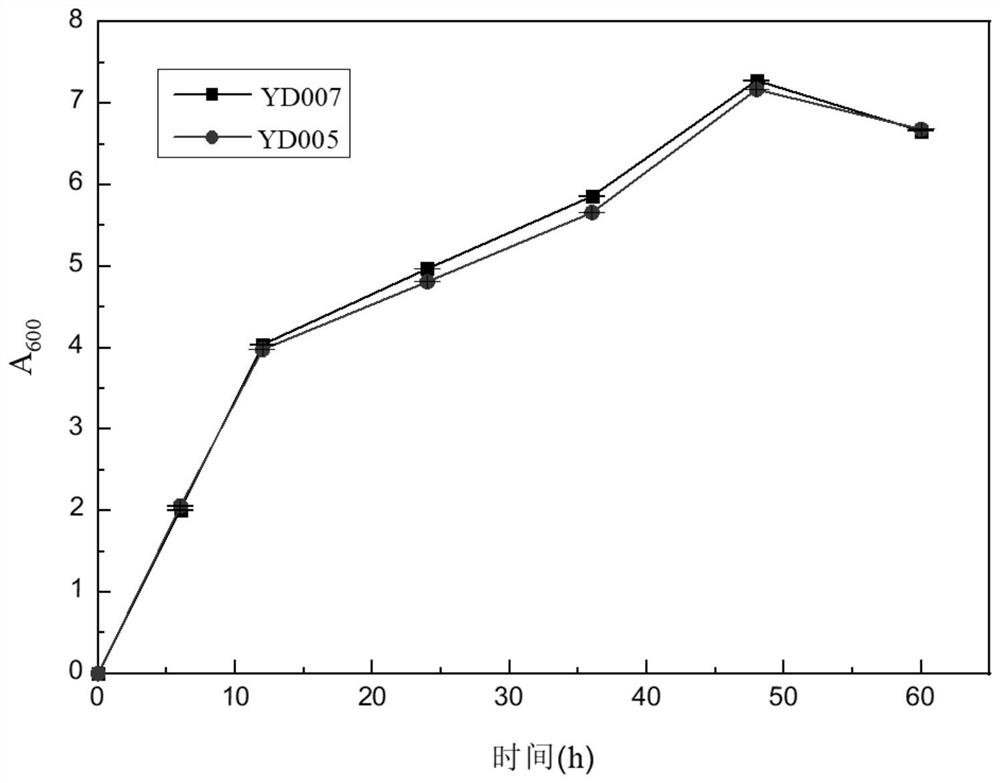
[0070] The gRNA expression cassette of GRE3, the gRNA expression cassette of PHO13, and the gRNA expression cassette of ASC1 constructed in Example 2, together with the p426-SNR52p-gRNA.CAN1.Y-SUP4t vector, XKS1 fragment, XLYA3 fragment, RPE1 fragment, and TAL1 fragment Yeast is transformed to knock out GRE3, PHO13, and ASC1 in the yeast genome, and at the same time introduce xylose metabolism pathways (XKS1 fragment, XLYA3 fragment, RPE1 fragment, TAL1 fragment). The CAN1 gene encodes arginine permease on the plasma membrane, and the p426-SNR52p-gRNA.CAN1.Y-SUP4t vector targets the CAN1 gene, because there is also a CAN1 homologous gene ALP1 on the genome, so even p426-SNR52p -gRNA.CAN1.Y-SUP4t vector edits CAN1 through non-homologous repair, and it will not affect the growth of yeast, which is also proved by experiments (such as image 3 shown). The content of the invention described in this ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com