Gene knockout method and its sgRNA fragment and application thereof
A gene knockout and fragment technology, applied in the field of gene editing, can solve the problems of complex operation and low editing efficiency, and achieve the effects of improving injection efficiency, ensuring editing efficiency, and simplifying vector construction technology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
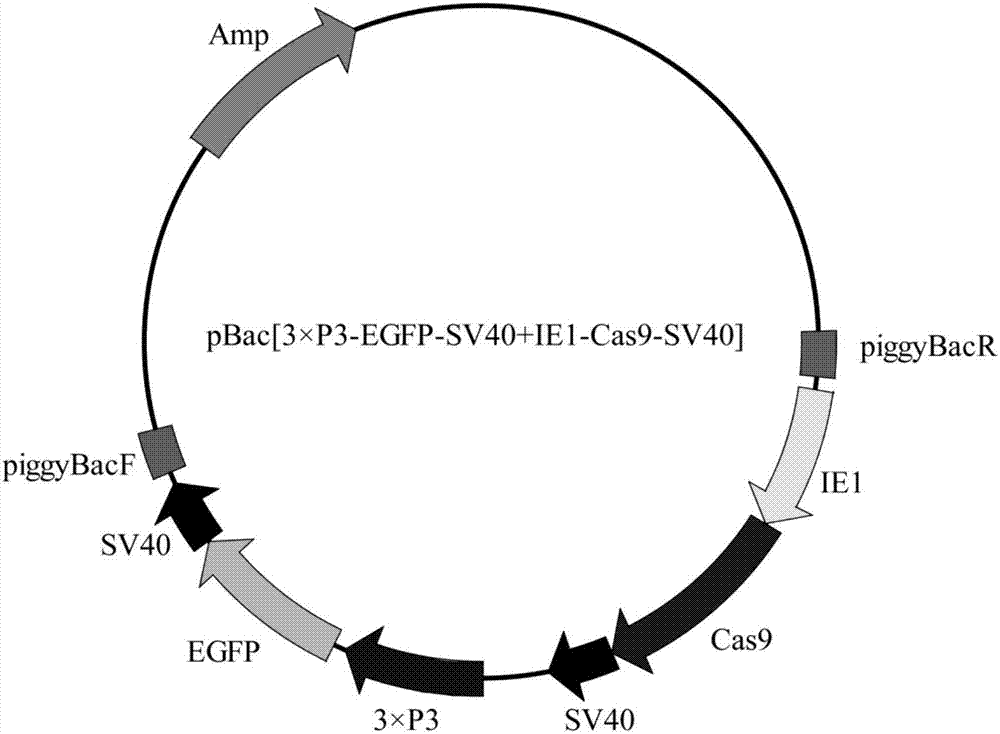
[0032] Construction of Cas9 expression vector pBac[3×p3-EGFP+IE1-Cas9-SV40] based on piggy Bac transposable vector.
[0033] figure 1 The schematic diagram of the expression vector pBac[3×p3-EGFP+IE1-Cas9-SV40] is shown.
[0034] The Cas9 gene comes from the vector pHsp70-Cas9 (purchased from ADDGENE, website: www.addgene.org / ), and the sequence of the Cas9 gene is shown in SEQ ID No.1.
[0035]Insert the Cas9 gene into the multiple cloning site of the starting vector pSL1180 to obtain an intermediate vector, and then insert the complete expression cassette IE1-Cas9-SV40 containing the Cas9 gene into the final vector pBac[3×P3-EGFP] to obtain a recombinant expression vector.
[0036] Specific operation method:
[0037] Step S1, preparation of vector pSL1180[Cas9-SV40]
[0038] The Cas9 gene was double-digested with the ClaI / XbaI restriction endonuclease carrier pHsp70-Cas9, and the Cas9 gene fragment was recovered (the recovery operation was performed according to the instr...
Embodiment 2
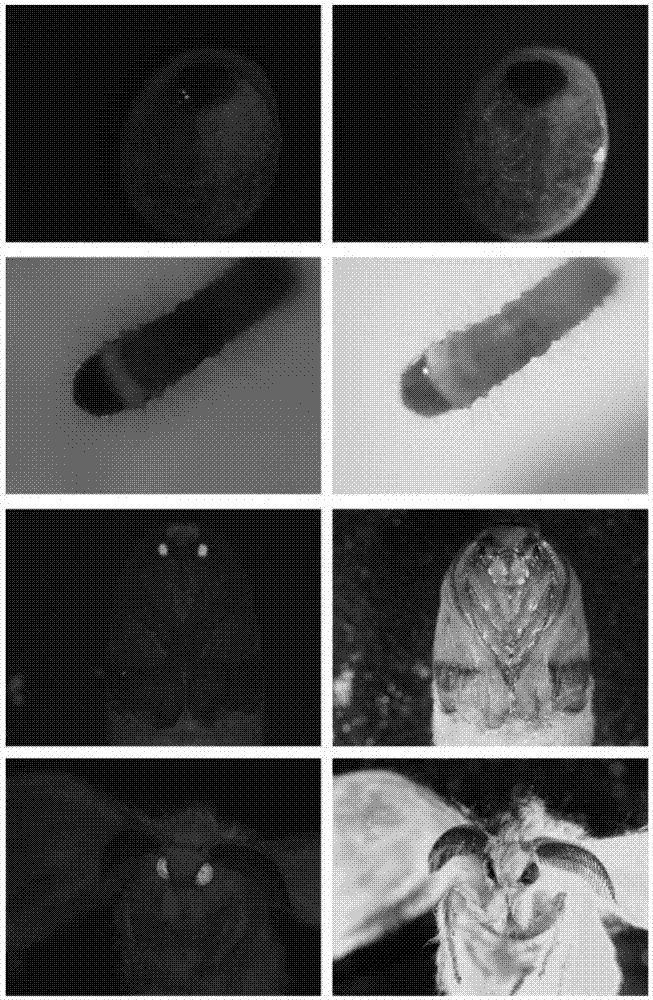
[0055] Breeding the transgenic line of silkworm with Cas9 gene.
[0056] The commercially available polymorphic silkworm line 305 was used as the raw material, and the eggs of the parental silkworms were reared on normal mulberry leaves and mated with moths to lay eggs.
[0057] Take 10nL-15nL of the recombinant vector plasmid pBac[3×P3-EGFP+IE1-Cas9-SV40] prepared in Example 1 with a concentration of 400ng / μL and mix it with the helper plasmid pHA3PIG. The mixed solution was injected into 400 silkworm eggs laid by female moths of silkworm strain 305 for 2 hours to 6 hours respectively, sealed with non-toxic glue and placed in an environment of 25°C and 85% relative humidity to accelerate hatching. After hatching, 156 G0 generation ant silkworms were obtained from pBac[3×P3-EGFP+IE1-Cas9-SV40].
[0058] The obtained ant silkworms were reared with mulberry leaves until they emerged as moths, and the obtained silkworm moths were backcrossed or self-crossed, and 37 moth circles ...
Embodiment 3
[0064] This example takes the silkworm endogenous fatty acid desaturase gene BmDES3 as an example, which is a member of the silkworm fatty acid desaturase gene family. It should be noted that the knockout method of the present invention is applicable to any Multiple knockout of a single endogenous gene of the target or a family of genes with similar domains. The sequence of the BmDES3 gene is shown in SEQID No.5.
[0065] Construction of targeting vector for endogenous fatty acid desaturase gene BmDES3 of silkworm.
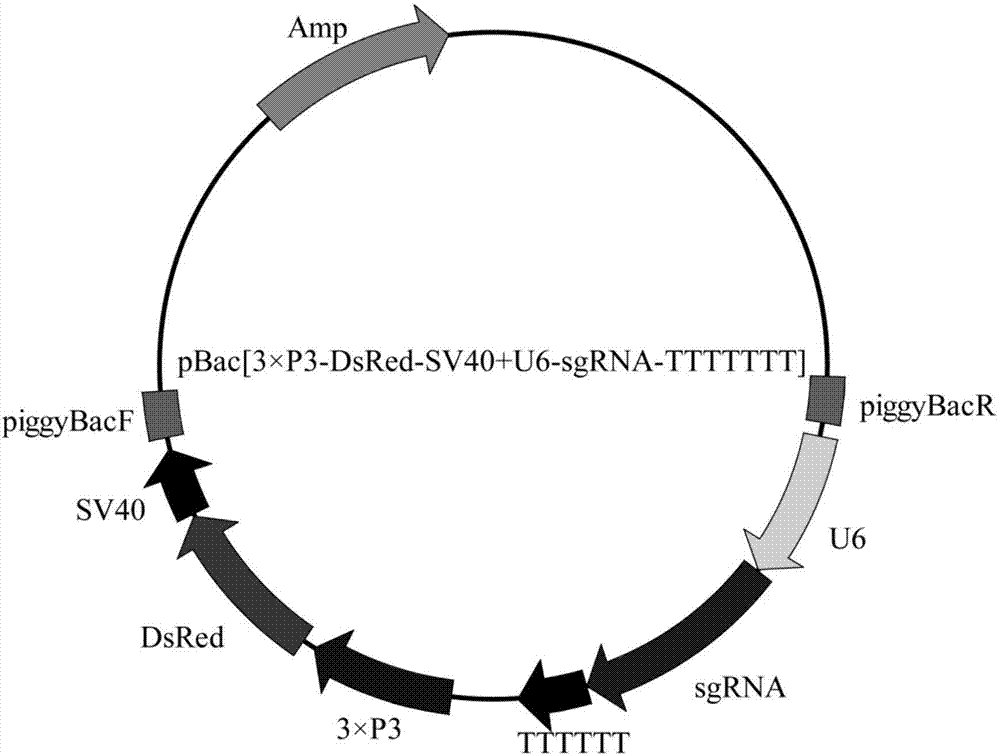
[0066] Insert the designed BmDES3-specific target sequence into the endogenous U6 promoter of silkworm, then connect it with sgRNA, and then insert the sgRNA expression cassette containing the complete target sequence into the final vector pBac[3×P3-DsRed] to obtain a recombinant expression vector .
[0067] figure 2 The schematic diagram of the expression vector pBac[3×P3-DsRed+U6-sgRNA] is shown.
[0068] Specific operation method:
[0069] Step S1, prepar...
PUM
| Property | Measurement | Unit |
|---|---|---|
| conversion efficiency | aaaaa | aaaaa |
| conversion efficiency | aaaaa | aaaaa |
| conversion efficiency | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com