High-throughput method for simultaneously detecting gene mutation and copy number change
A high-throughput, copy-number technology, applied in the field of biomedicine, can solve problems such as simultaneous detection, and achieve the effects of improved accuracy, reduced detection cost, and improved detection throughput
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
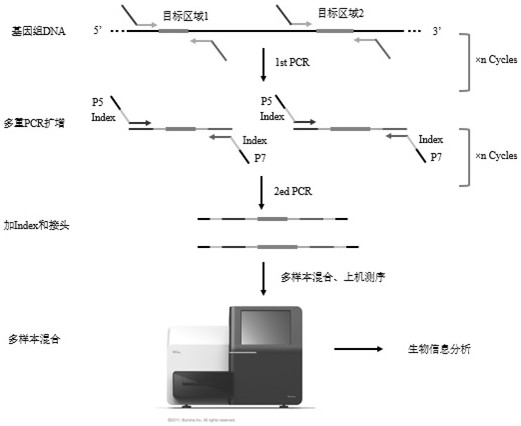
[0021] Example as figure 1 As shown, a high-throughput method for simultaneously detecting gene mutations and copy number changes includes: designing a pair of positive-strand probes and negative-strand probes for multiple sites or regions to be detected, and each pair of positive-strand The positive strand probe in the probe and the negative strand probe is located on the positive strand of the genome sequence, and the negative strand probe in each pair of positive strand probe and negative strand probe is located on the negative strand of the genome sequence; The length of the strand probe or negative strand probe is 18-36bp, preferably 20-27bp.
[0022] Use the positive-strand probe and the negative-strand probe to combine to obtain a combined probe, and perform the first round of PCR amplification on the DNA to be tested based on the combined probe; for the product obtained by the first round of PCR amplification, use a Amplify the universal amplification primers that mat...
Embodiment 2
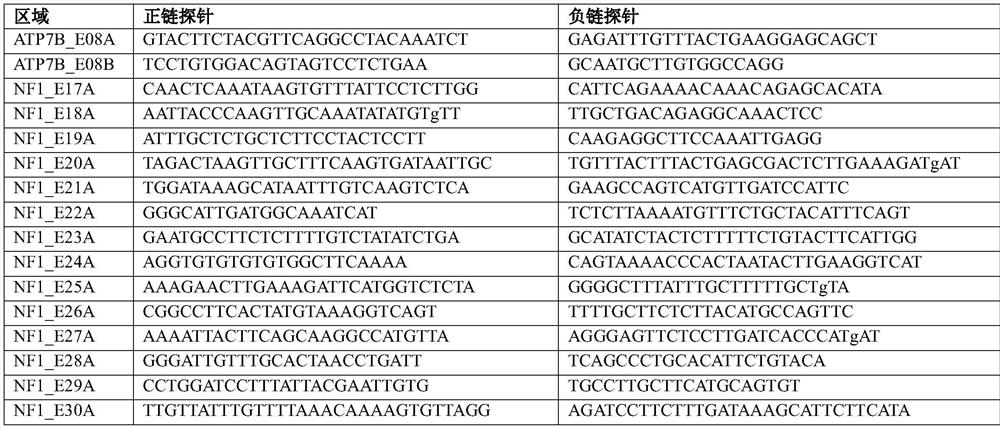
[0025] Example 2 Design probes for the coding regions of 4 genes (ATP7B, NF1, TSC1, TSC2), the probes and general primer information are shown in Table 1:
[0026] Table 1
[0027]
[0028]
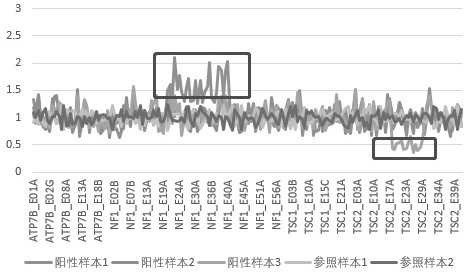
[0029] The three positive samples of the known mutant type were amplified by PCR using a mixture of positive-strand probes and negative-strand probes at each site to obtain amplified products; the amplified products of the two panels were mixed, and then universally used with illumina with different tag sequences PCR primers compatible with the sequencing platform were used for amplification, and the products of each sample were uniformly mixed and then sequenced on an illumina sequencing instrument in PE150 mode, and the sequencing data were subsequently analyzed.
[0030] Experimental procedure: (1) Quantify the concentration of wild-type samples and mutant samples; (2) Configure two panels of positive-strand probe and negative-strand probe mixture, each primer concentration is 2uM;...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com