SARS-CoV-2 virus S protein receptor binding region encoding gene, antibody and application thereof
A receptor binding, sars-cov-2 technology, applied in the field of biomedicine, to achieve the effect of ensuring uniqueness, simple operation and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
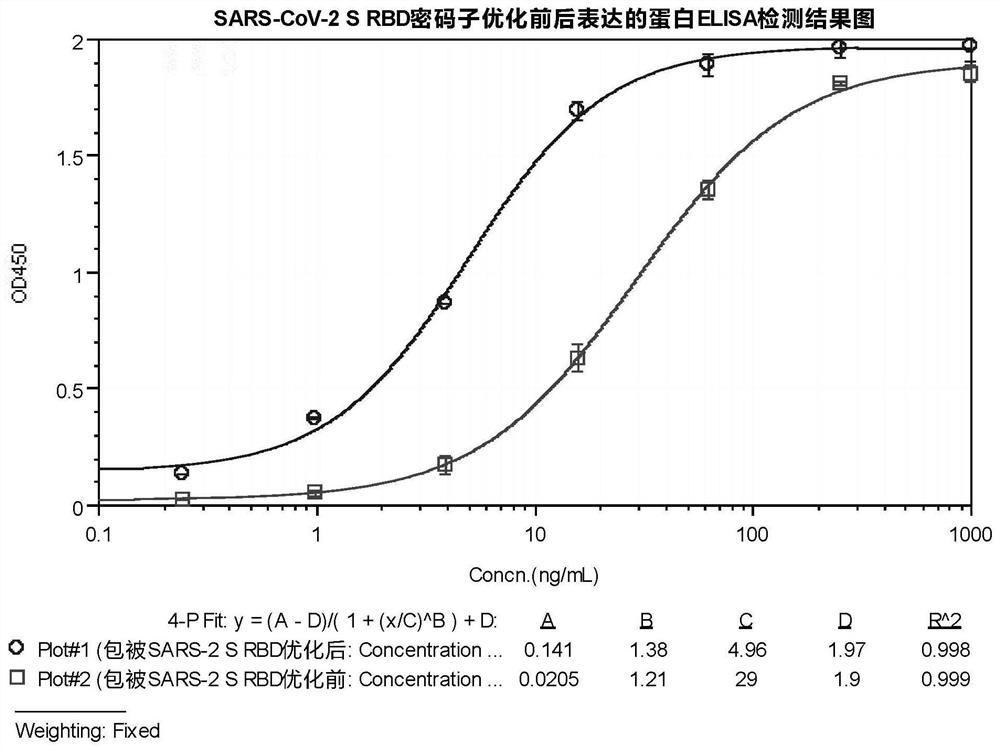
[0068] Example 1 SARS-CoV-2 S RBD coding gene optimization and expression
[0069] The amino acid sequence position of the sugar spike protein S RBD (Spike protein RBD, S RBD) of SARS-CoV-2 is aa319-aa541, as shown in UniProt-PODTC2 (SPIKE_SARS2) for details, expressing the SARS-CoV-2 spike protein S The native nucleotide sequence of RBD is as follows (SEQ ID NO: 1).
[0070] AGAGTCCAACCAACAGAATCTATTGTTAGATTTCCTAATATTACAAA CTTGTGCCCTTTTGGTGAAGTTTTTAACGCCACCAGATTTGCATCTGTTTA TGCTTGGAACAGGAAGAGAATCAGCAACTGTGTTGCTGATTATTCTGTCC TATATAATTCCGCATCATTTTCCACTTTTAAGTGTTATGGAGTGTCTCCTA CTAAATTAAATGATCTCTGCTTTACTAATGTCTATGCAGATTCATTTGTAA TTAGAGGTGATGAAGTCAGACAAATCGCTCCAGGGCAAACTGGAAAGAT TGCTGATTATAATTATAAATTACCAGATGATTTTACAGGCTGCGTTATAGC TTGGAATTCTAACAATCTTGATTCTAAGGTTGGTGGTAATTATAATTACCT GTATAGATTGTTTAGGAAGTCTAATCTCAAACCTTTTGAGAGAGATATTT CAACTGAAATCTATCAGGCCGGTAGCACACCTTGTAATGGTGTTGAAGGT TTTAATTGTTACTTTCCTTTACAATCATATGGTTTCCAACCCACTAATGGT GTTGGTTACCAACCATACAGAGTAGTAGTACTTTCTTTTGAACTTCTACA...
Embodiment 2
[0077] Example 2 Expression and purification of SARS-Cov-2 S RBD recombinant protein
[0078] The C-terminus of the expressed protein of SEQ ID NO:2 is linked to the Fc fragment of human IgG1 (P01857) for purification and subsequent detection.
[0079] Synthesize the BRP2 gene fragment, connect the BRP2 gene fragment to the upper pBV vector (Bailing Bio), and transform it into Escherichia coli for 16 hours at 37°C.
[0080] The next day, the BRP2 plasmid was lysed and extracted. According to a conventional method, 50 μg of pBV-BRP2 was used to transfect 50 mL of 293-6E cells with cationic liposomes. After 6 days of transfection, the cells and supernatant were collected, and the collected cells were lysed to prepare a cell lysate. The supernatant after transfection was collected, the recombinant protein was purified using a nickel column, and the buffer was replaced with PBS. Purified samples were stored at 4°C.
Embodiment 3
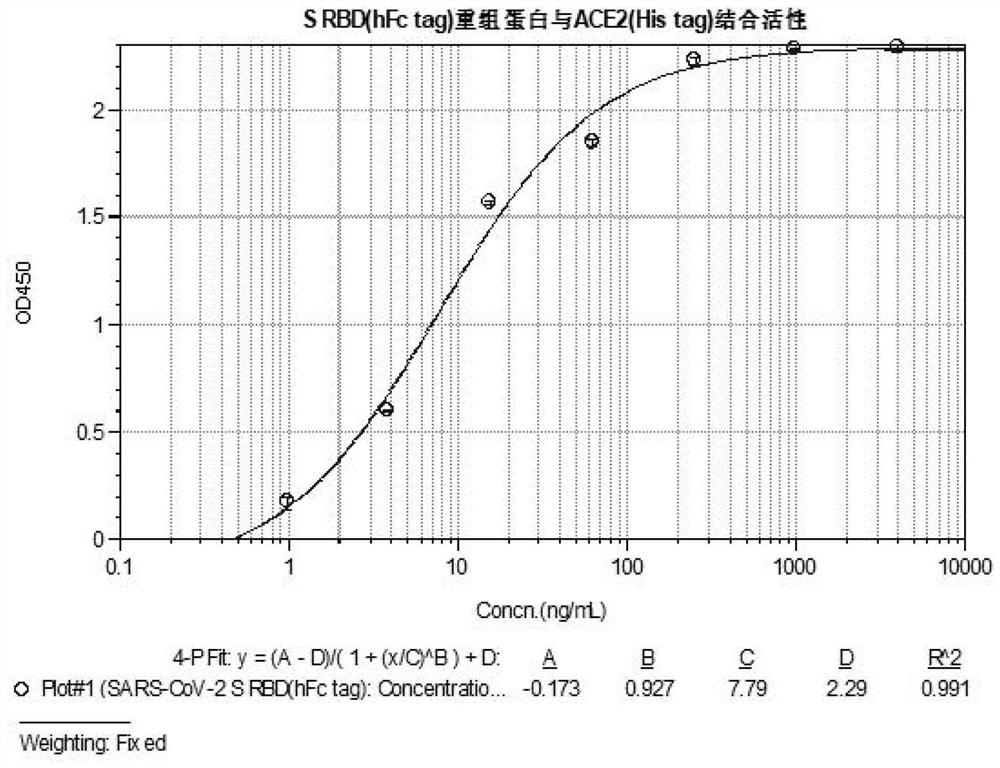
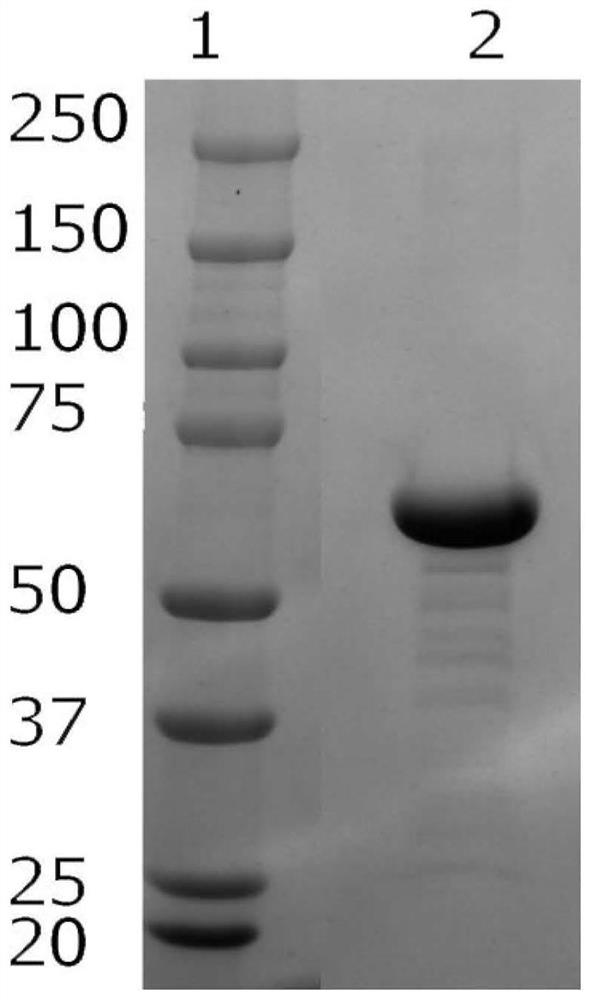
[0081] Example 3 Identification of SARS-CoV-2 S RBD recombinant protein
[0082] The SARS-CoV-2 S RBD recombinant protein was identified using ELISA, SDS-PAGE, and Western blot respectively. The specific identification methods are as follows:
[0083] 1. Identification of ELISA method
[0084] 1) Coat the ACE2 recombinant protein (with His tag) receptor in the extracellular region (Cat. No.: ACE-HM401, Shanghai Kaijia Biology), dilute the ACE2 recombinant protein concentration in carbonate buffer to 2 μg / mL, 50 μL / well ELISA plate was coated overnight at 4°C.
[0085] 2) The next day, use 0.01M, pH7.4 PBST to place the coated microplate plate on a plate washer and wash the plate 3 times, 250 μL / well.
[0086] 3) Add 100 μL of 3% BSA / PBST solution to each well, block and incubate at 30°C for 1h-2h.
[0087] 4) After washing the plate, add SARS-CoV-2 S RBD recombinant protein ligand starting from 4 μg / mL and 4-fold dilution to each well, 50 μL / well. Incubate at 30°C for abou...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com