Application of non-coding gene capable of controlling corn ear thickness heterosis and precursor sequence of non-coding gene
A heterosis, non-coding technology, applied in the application of non-coding genes and their precursor sequences, can solve the problems of low probability, poor predictability, and large blindness of screening excellent combinations, and achieve the improvement of ear diameter and ear size. yield effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Discovery of maize ear coarse heterosis non-coding gene zma-miR390 involved in the determination of maize ear heterosis:
[0026] A set of single-segment substitution line populations was constructed with the excellent maize inbred line Chang 7-2 as the donor parent and lx9801 as the recipient parent; the corresponding testcross population was constructed by crossing with the testcross parent Zheng 58 (Zheng 58× SSSL); By comparing the phenotypic differences between the test cross and the control Zheng 58×lx9801 (Ludan 9002), a single-segment substitution line containing the heterosis locus of panicle diameter was identified and named lx9801 hlEW2b ;For Zheng 58×lx9801, Zheng 58×lx9801 hlEW2b Ears at different developmental stages were subjected to transcriptome sequencing, degradome sequencing and miRNA-seq;
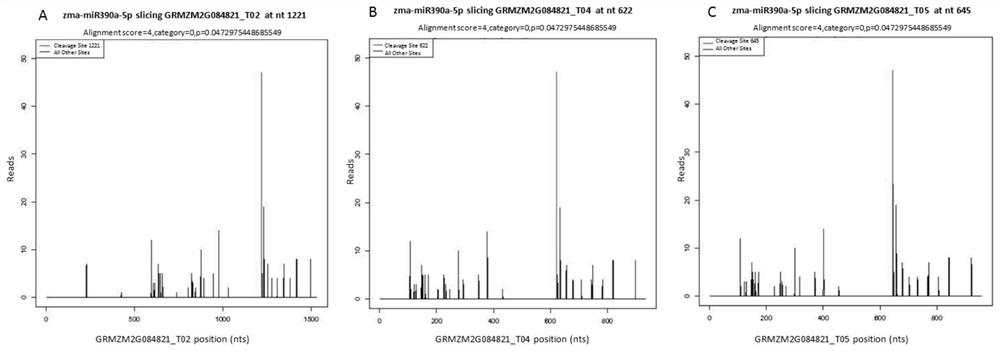
[0027] The results of miRNA-seq showed that there were significant differences in the expression of zma-miR390 between the two materials; the results of maize e...
Embodiment 2
[0031] Confirmation of maize ear coarse heterosis non-coding gene zma-miR390 regulating maize ear heterosis:
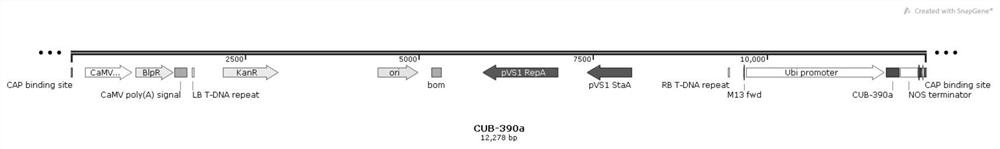
[0032] The pCAMBIA3301 vector was digested with Bgl II and BstE II enzymes, and then the artificially synthesized Zma-miR390a fragment was connected to the pCAMBIA3301 vector using T4 DNA ligase to construct an overexpression vector of zma-miR390a (pCAMBIA3301::35S::Zma-miR390a , figure 2), the RNA sequence of the Zma-miR390a gene is shown in SEQ ID NO.2, and the primers used to amplify Zma-miR390a include CUB-mir390a-F primers and CUB-mir390a-R primers, and the CUB-mir390a-F primers The DNA sequence of the CUB-mir390a-R primer is shown in SEQ ID NO.3, the DNA sequence of the CUB-mir390a-R primer is shown in SEQ ID NO.4, and the DNA sequence of the overexpression vector pCAMBIA3301::35S::Zma-miR390a is shown in SEQ ID NO.4 As shown in ID NO.5;
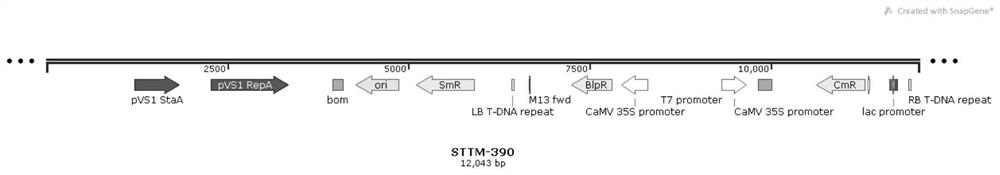
[0033] With the sequence of zma-miR390 as the target, construct the STTM390 vector (such as image 3 ), the DNA sequence...
Embodiment 3
[0036] Evolutionary conservation of the non-coding gene zma-miR390 in maize ear coarse heterosis:
[0037] MiR390 is an ancient and highly conserved miRNAs. At present, most of the research on plant miR390 genes is focused on model plants such as Arabidopsis and rice, and high-throughput sequencing technology combined with bioinformatics is often used to identify target genes and action sites. Point and function forecasting and more. Download the precursor sequences of gene family members of maize, Arabidopsis, rice, and soybean miR390 from miRBase (http: / / www.mirbase.org / ), and use the ClustalW method of MEGAX software to compare the precursor sequences of miR390 Yes, choose the Neighbor-Joining method to construct the phylogenetic tree (such as Figure 6 ).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com