Method and kit for organism fusion gene detection and fusion abundance quantification
A technology of fusion genes and organisms, which can be used in biochemical equipment and methods, and the determination/inspection of microorganisms. , the effect of reducing the risk of opening tubes / pipetting
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
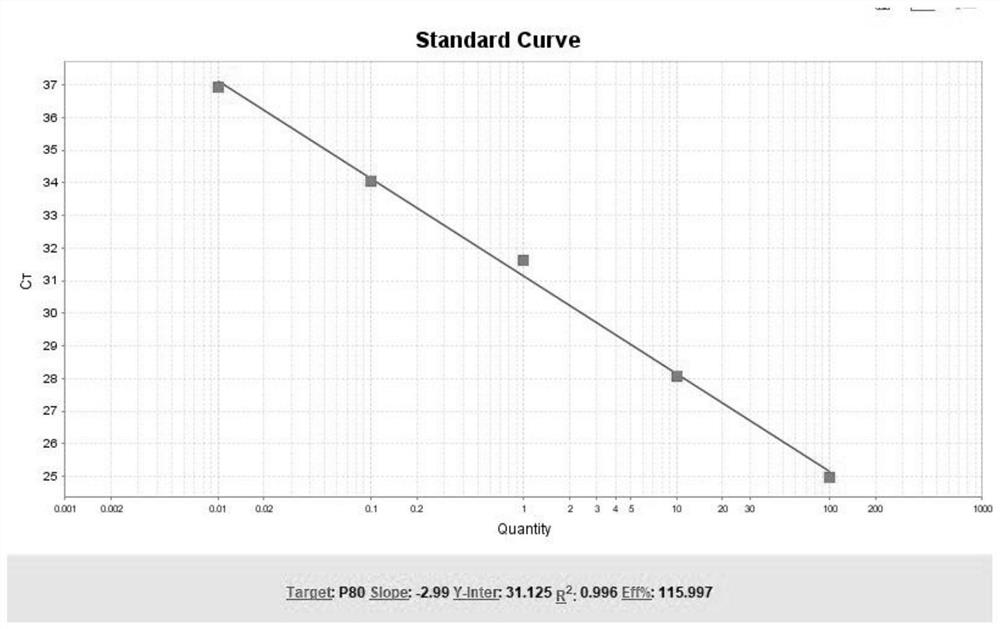
[0045] Example 1 The primer sequence for amplifying tumor-related gene mutation sites was synthesized by Shanghai Sangong, a primer synthesis company.
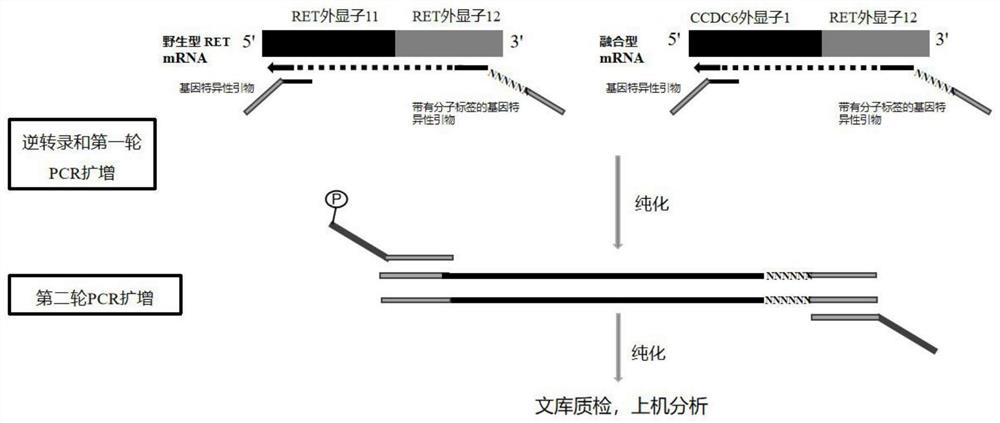
[0046] Taking CCDC6-RET as an example, the RNA primer design principle of the present invention will be described in detail below. Such as figure 1 As shown, with the position of the breakpoint as the boundary, exon 1 of the 5' end CCDC6 gene is fused with exon 12 of the 3' RET gene, so the upstream primer is designed on exon 1 of the CCDC6 gene, and the downstream primer is designed on the RET gene On exon 12 of the gene; the RET wild-type upstream primer was designed on exon 11, and the downstream was consistent with the downstream primer of the fusion gene.
[0047] (1) RNA-specific primers, the primer sequences of which are shown in Table 2:
[0048] Table 2 fusion gene and its primer sequence
[0049]
[0050]
[0051] Wild-type gene and its primer sequence
[0052]
[0053]
[0054] *P stands for phosphor...
Embodiment 2
[0061] Example 2: Sample pretreatment and nucleic acid extraction
[0062] Qualified professionals take samples with a puncture needle, and the samples include puncture tissue samples greater than 1 mg. After the biopsy is completed, infiltrate the tissue completely in RNA preservation solution as soon as possible. Samples were stored at -20°C prior to sample shipment.
[0063] Use the tissue cell RNA extraction kit (spin column method) and refer to the instructions of the kit to extract the RNA in the sample.
[0064] 1. Mechanical tissue sample homogenization:
[0065] Put the sample in a suitable glass tube or centrifuge tube, add 350 μL Buffer RL and DTT mixed lysate (100:1) lysate, insert the probe into the lysate, and homogenize intermittently at high speed, each time for 15-20 seconds until the sample Homogenize completely.
[0066] 2. Extraction of nucleic acid after homogenization
[0067] About 350 μL of the homogenized sample was centrifuged at 14000 rpm for 3 ...
Embodiment 3
[0078] Embodiment 3: Construction of library
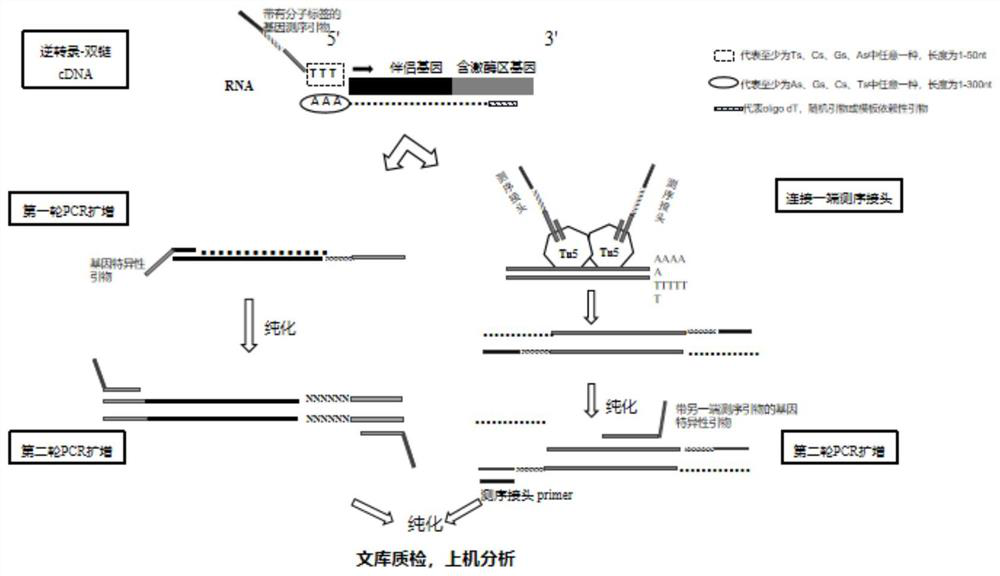
[0079] The library construction process of this kit is as follows:
[0080] (1) RNA reverse transcription and the first round of PCR amplification (amplification of the target region) uses RNA as a template, and the reverse primer of the fusion 3' end gene with 6 random bases is used as a specific primer for reverse transcription to obtain cDNA ; Use specific multiple PCR primers and amplification reagents to amplify cDNA to obtain specific target fragments, and the reaction is carried out in a single tube. Vazyme's reverse transcriptase and its KAPA multi-reconstruction library Mix were used to prepare the following system:
[0081]
[0082] Set the following conditions on the PCR instrument for the reaction:
[0083]
[0084] (2) The first round of PCR product purification
[0085] 1) After the programmed reaction, the sample was briefly centrifuged, and the sample was added with nucleic acid-free water to make up 20 μL, ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com