Visual nucleic acid detection method for simultaneously detecting one or plurality of kinds of target nucleic acids and application of visual nucleic acid detection method
A technology of target nucleic acid and detection method, applied in the field of molecular biology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0077] Embodiment 1: experimental material and method steps of the present invention
[0078] The materials and primer sequences used in the present invention are shown in Table 1 and Table 2; wherein, the DNA and RNA were synthesized by Sangon Bioengineering (Shanghai) Co., Ltd.
[0079] Among them, Bst DNA polymerase (M0275S), Nb.BbvCI (R0631S), Nb.BtsI (R0707S), 10Xbuffer 1.1 (100mM Bis-Tris-HCl pH7, 100mM MgCl 2 , 1mg / mL BSA) etc. were purchased from NEB. HybriDetect test strip detection kit (MGHD 1, MGHD 2) was purchased from Milenia Biotec GmbH (Germany).
[0080] Primers and sequences used in Table 1
[0081]
[0082] *Represents thio modification
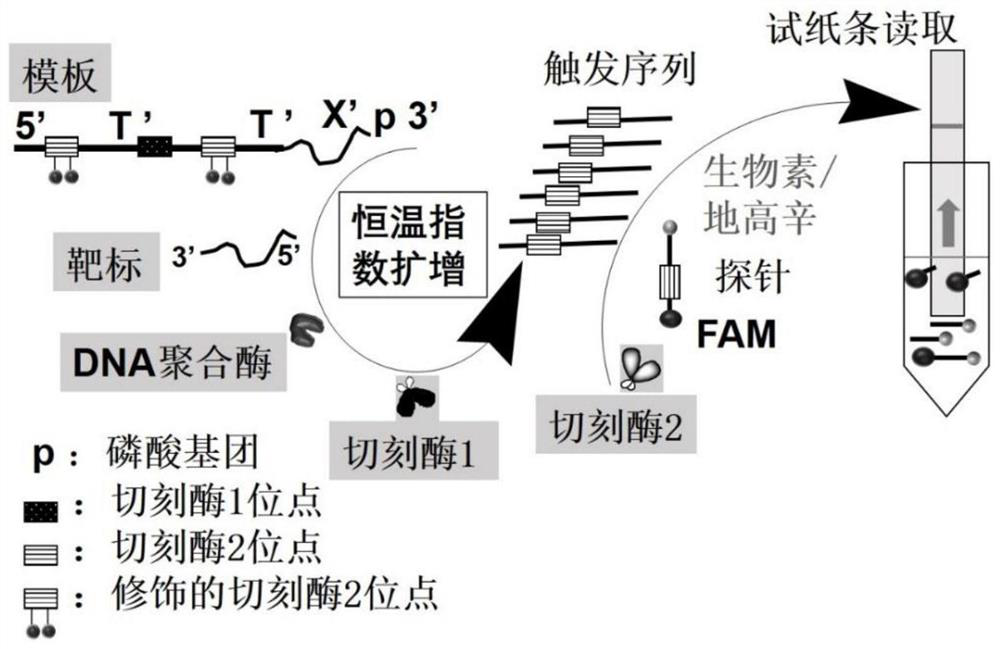
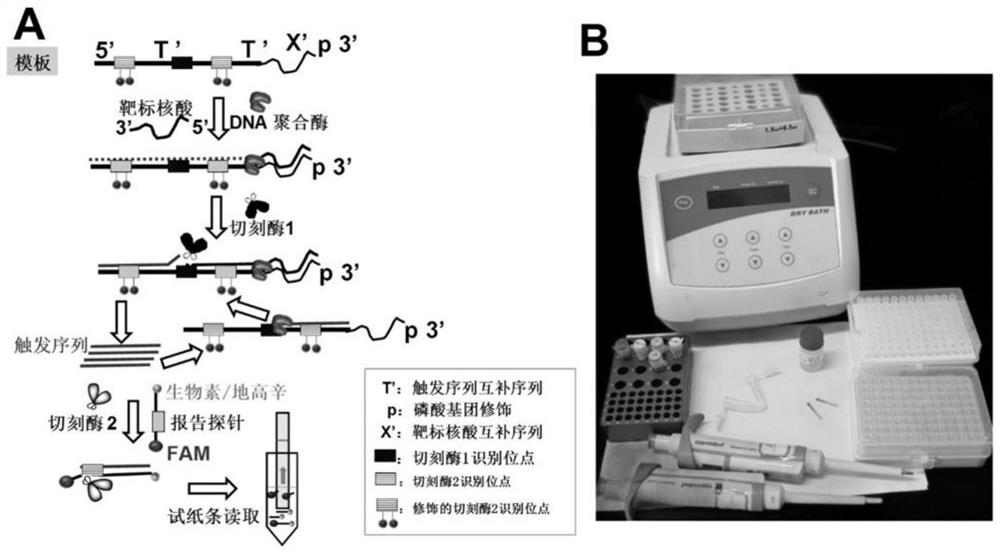
[0083] combine figure 1 and figure 2 Described in detail the exemplary experimental steps of the present invention, specifically as follows:
[0084] 1. The primers are dissolved in DEPC water, and then the OD260 value is measured by nanodrop, and the primers are calculated according to the formula A=εbc (A is th...
Embodiment 2
[0094] Embodiment 2: method principle verification of the present invention
[0095] according to figure 1 Principle, the template DNA is Pre-TrigTel, as shown in SEQ ID NO: 1, wherein the 3' end sequence is: CTAACCCTAACCCTAACCCTAA. When configuring the reaction sample, add the following components in order:
[0096] Table 4 Reagents in tube A reaction system
[0097] Added reagent μL / reaction 10X Buffer 1.1 0.5 100nM TrigTel 1 10 μM MB-TrigTel 1 2.5mM dNTPs 1 100nM TS-3 1
[0098] Table 5 Reagents in tube B reaction system
[0099] Added reagent μL / reaction 10X Buffer 1.1 0.5 10U / μL Nb.BbvCI 0.25 8U / μL Bst DNA polymerase 0.125 10U / μL Nb.BtsI 0.25 500ng / μL ET SSB 0.1 water 4.275
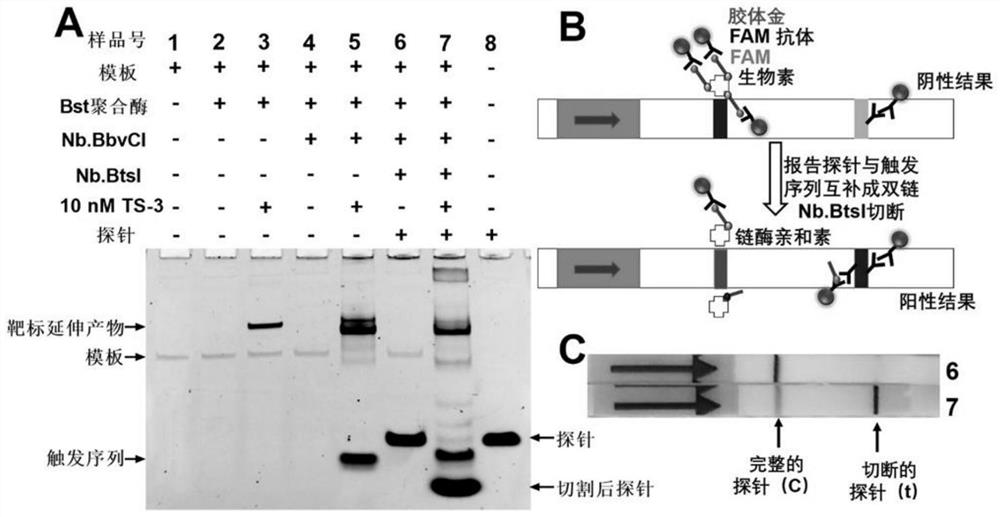
[0100] The total volume of each reaction is 10 μL, and the sample is kept at about 4 °C during the preparation of the sample. In order to verify the principle of the scheme of the present inventio...
Embodiment 3
[0101] Example 3: Detection of telomerase extension products using the method of the present invention
[0102] according to figure 1 Principle, the template DNA is Pre-TrigTel, as shown in SEQ ID NO: 1, wherein the 3' end sequence is: CTAACCCTAACCCTAACCCTAA. When configuring the reaction sample, add the following components in order:
[0103] Table 6 Reagents in tube A reaction system
[0104] Added reagent μL / reaction 10X Buffer 1.1 0.5 100nM TrigTel 1 10 μM MB-TrigTel 1 2.5mM dNTPs 1 TS / TS-1 / TS-2 / TS-3 / TS-4 1
[0105] Table 7 Reagents in tube B reaction system
[0106] Added reagent μL / reaction 10X Buffer 1.1 0.5 10U / μL Nb.BbvCI 0.25 8U / μL Bst DNA polymerase 0.125 10U / μL Nb.BtsI 0.25 500ng / μL ET SSB 0.1 water 4.275
[0107] The total volume of each reaction is 10 μL, and the sample is kept at about 4 °C during the preparation of the sample. In the test for the detection o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com