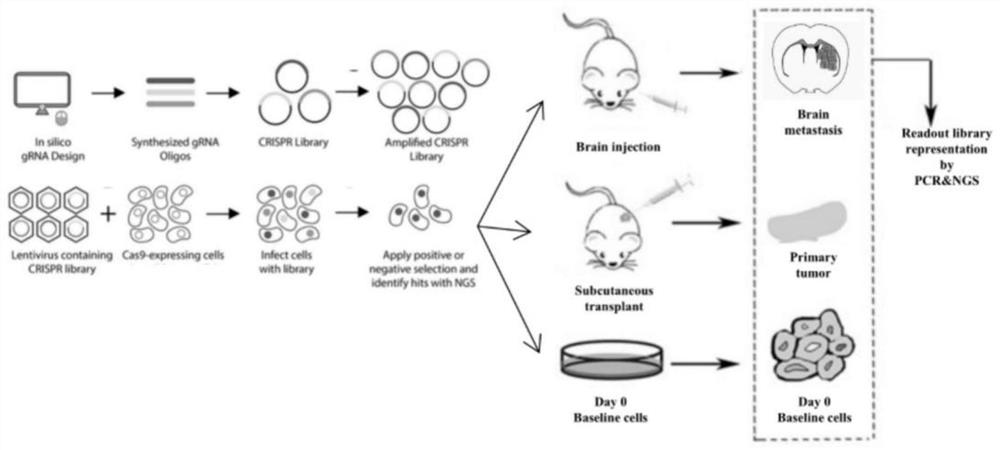
CRISPR/Cas9 library high-throughput screening method for brain metastasis related genes
A screening method and technology for brain metastases, applied in the field of cancer diagnosis, can solve problems that have not been raised by researchers
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Paired samples of breast cancer primary tumors and brain metastases were collected from 5 triple-negative breast cancer patients with simple brain metastases, a total of 5 pairs of 10 samples. Use the RNA-Quick Purification Kit (ES Science #RN001) kit to extract sample RNA and perform RNA-Seq detection. The detection results of RNA-seq were analyzed, and the Fold change value was greater than 1.5 at the same time as the screening condition to screen differentially expressed genes, and a total of 597 highly expressed genes in brain metastases were obtained (as shown in Table 1).
[0038] Table 1 Genes associated with brain metastasis of triple-negative breast cancer
[0039]
[0040]
Embodiment 2
[0042] According to the sgRNA online design software (http: / / crispr.mit.edu / ), 6 sgRNAs were designed for each gene in Example 1. Each sgRNA is 20bp, and a 27bp homology arm is added on the left and right sides of the sgRNA (the sequence of the left homology arm is ATATCTTGTGGAAAGGACGAAACACCG (SEQ ID NO: 1), and the sequence of the right homology arm is GTTTTAGAGCTAGAAATAGCAAGTTAA (SEQ ID NO :2)), for example figure 2 shown. In situ synthesis of oligonucleotide probe pools targeting multiple genes on a high-throughput chip.
Embodiment 3
[0044] 1. Amplify the oligonucleotide probe pool
[0045] Add TE Buffer to the powder of the oligonucleotide probe pool to resuspend, and use the following reaction system for polymerase chain reaction (polymerase chain reaction, PCR):
[0046] The PCR amplification program is:
[0047]
[0048] The PCR amplification system is:
[0049]
[0050] Primers are:
[0051] A. Forward primer:
[0052] GTAACTTGAAAGTATTTCGATTTCTTGGCTTTATATATCTTGTGGAAAGGACGAAACACC (SEQ ID NO: 3)
[0053] B. Reverse primer:
[0054] ACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC (SEQ ID NO: 4)
[0055] 2. Digest the lentiGuide-puro vector
[0057]
[0058] Enzyme digestion system temperature setting:
[0059] Step1: 90 minutes at 55°C
[0060] Step2: 4°C°
[0061] The digested product was subjected to DNA electrophoresis on 2% agarose gel, and the 10,000 bp band was excised and recovered by Fastpuregel DNA Extraction Mini Kit (Novizyme #DC301...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com