Nitrile hydratase lysine mutant HBA-K2H1, coding gene and application
A nitrile hydratase lysine, encoding gene technology, applied in the application, genetic engineering, plant genetic improvement and other directions, can solve problems such as artificial regulation of the self-modification efficiency of nitrile hydratase
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Example 1: Site-directed mutagenesis of nitrile hydratase HBA
[0038] 1) Extract the recombinant plasmid pET-28-HBA containing the nitrile hydratase HBA gene from recombinant Escherichia coli according to the instructions of the plasmid mini-extraction kit from Axygen Company. The amino acid sequence of wild-type nitrile hydratase HBA is shown in SEQ ID NO.3.
[0039] 2) Design site-directed mutagenesis primers 5'-CAAACATGGTTTCGGCAAGATCATTCGTGAGAAAAACGAGCCGCTGTTCC-3'(SEQ ID NO.5) and 5'-GATCTTGCCGAAACCATGTTTGCCACCCAGG TCGTGG-3'(SEQ ID NO.5) according to the nucleotide sequence (SEQ ID NO.4) of wild-type nitrile hydratase HBA ID NO.6).
[0040] 3) According to the instructions of the Fast Mutagenesis System Kit of Beijing Quanshijin Biotechnology Co., Ltd., the recombinant plasmid pET-28-HBA obtained in step 1 was used as a template, and the primers synthesized in step 2 were used for single-site site-directed mutagenesis PCR.
[0041] 4) Transform the site-directed m...
Embodiment 2
[0042] Embodiment 2: Enzyme preparation of nitrile hydratase mutant HBA-K2H1 and wild-type HBA
[0043] 1) Recombinant strains containing mutant HBA-K2H1 and wild-type HBA were respectively inoculated in LB (50 μg / mL Kan) culture medium at an inoculation amount of 0.1%, and cultured with shaking at 180 rpm for 16 hours at 37°C.
[0044] 2) Inoculate the 1% inoculum of the activated bacterial solution overnight into fresh LB (containing 50 μg / mL Kan) culture solution, culture at 37°C with shaking at 180 rpm for 3 h (OD 600nm reach 0.6-1.0).
[0045] 3) Add IPTG (isopropylthiogalactopyranoside) at a final concentration of 0.5 mM for induction, and culture at 20° C. with shaking at 150 rpm for about 20 h.
[0046] 4) Centrifuge at 8000rpm for 5min, collect the cells and suspend the cells with PBS buffer.
[0047] 5) Ultrasonic disrupt the bacteria in a low-temperature ice-water bath. Ultrasonic crushing conditions are: power 300W, 5s on, 10s off, temperature 4°C, effective cru...
Embodiment 3
[0050] Embodiment 3: the property determination of the purified enzyme of nitrile hydratase mutant HBA-K2H1 and wild-type HBA
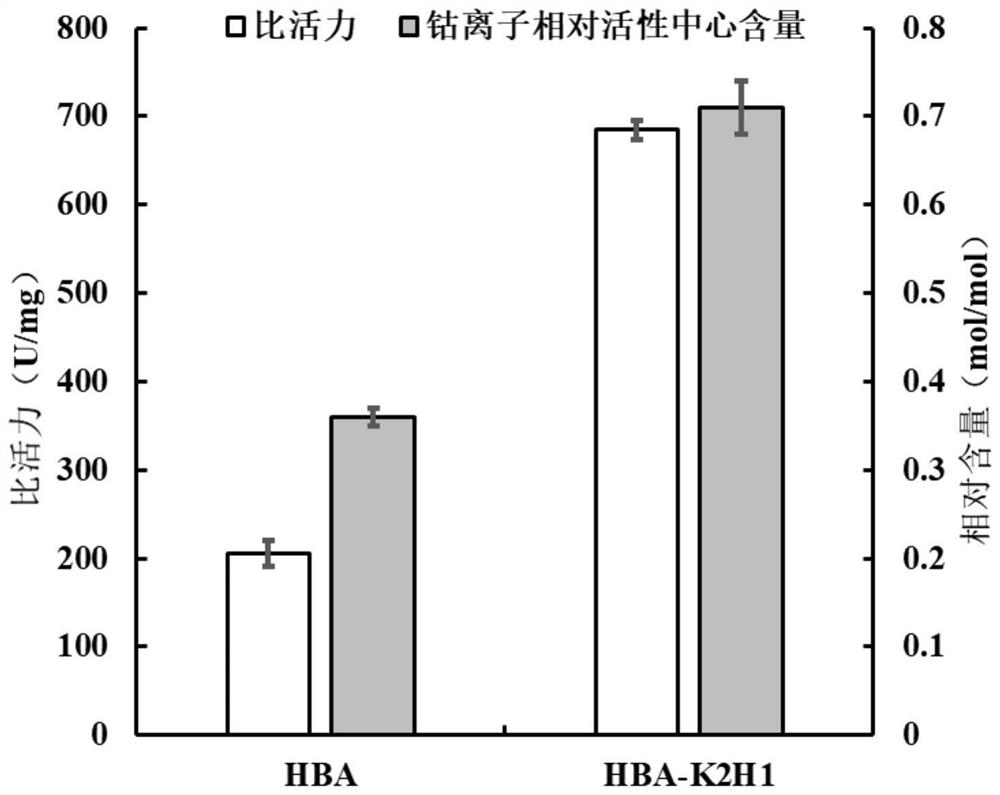
[0051] 1) Activity analysis of mutant HBA-K2H1 and wild-type HBA purified enzymes
[0052] Dissolve the substrate 3-cyanopyridine in 50mM boric acid-borax buffer solution (pH 8.0) to a final concentration of 10mM. After preheating the reaction solution at 40°C for 5min, add the enzyme solution to a final concentration of 3.2μg / mL, react at 40°C for 10min, then add 5M concentrated HCl
[0053] 20 μL to stop the reaction. After removing impurities by filtration with a 0.22 μm filter membrane, the amount of the product 3-pyridinecarboxamide formed was detected by HPLC. The detection condition of HPLC is to use Agilent InfinityLab Poroshell 120EC-C18 column (4.6×150mm, 4μm) chromatographic column, the column temperature is 36°C, the mobile phase is 10% (v / v) acetonitrile, and the flow rate is 0.5mL / min. The wavelength is 215nm; 1 enzyme activity unit (U...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com