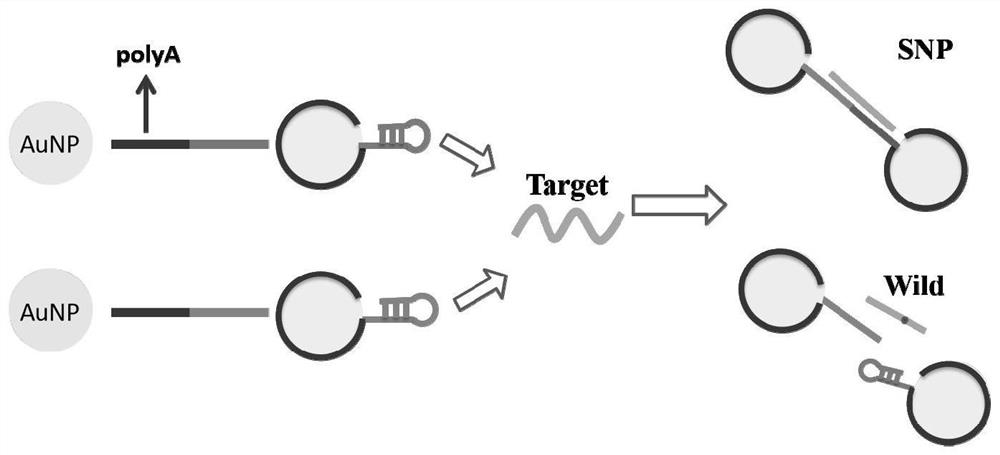
Detection probe for SNP site detection, detection method and application of detection probe
A technology for detecting probes and sites, applied in biochemical equipment and methods, recombinant DNA technology, microbial measurement/inspection, etc., to achieve the effects of fast hole passing, improved detection resolution, and reduced translocation speed
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0061] Embodiment 1: the preparation of detection probe
[0062] Step 1: Take an appropriate amount of the first nucleic acid probe (SEQ ID NO.1) and gold nanospheres with a particle size of about 5nm, mix the DNA solution and the gold nanospheres according to the ratio range (molar ratio) of 1:10, shake well, Cultivate overnight in an incubator at 25°C and 300 rpm.
[0063] Step 2: Take the PBS buffer solution with a volume of about 10% of the mixed solution in step (1), so that the PBS concentration in the final solution is about 0.1mol / L. The PBS buffer solution is divided into 5 times with an interval of 20 minutes each time Add, fully shake and shake well after each addition, after all the addition, incubate overnight in an incubator at 25°C and 300rpm:
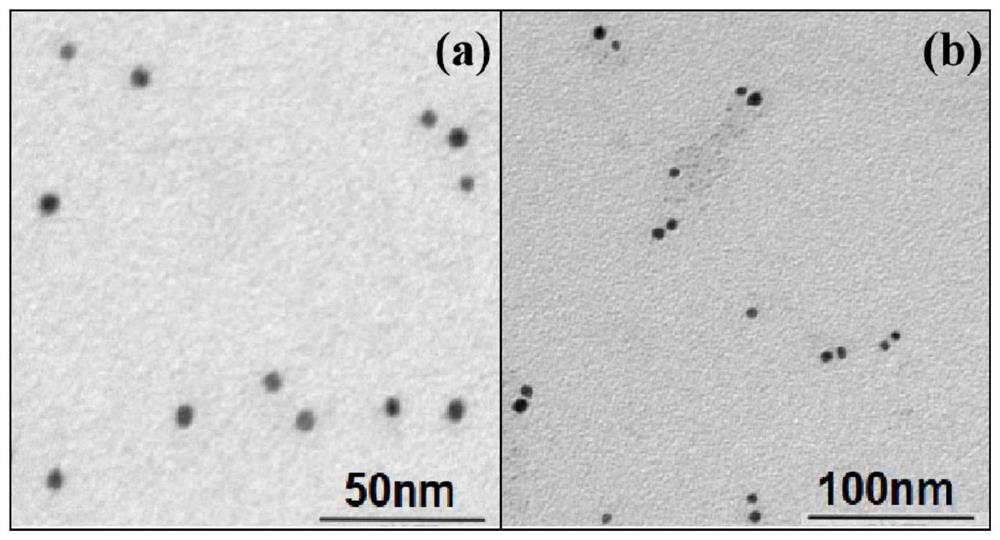
[0064] Step 3: Place the sample in a high-speed temperature-controlled centrifuge, conditions: 15,000rpm, 25°C, after high-speed centrifugation, a precipitate appears at the bottom, that is, gold nanospheres modified wi...
Embodiment 2
[0066] Embodiment 2: nanopore sensor detection
[0067] In this embodiment, the present invention selects the wild-type and mutant DNA fragments in the p53 gene as detection objects.
[0068] Step 1: Take the two probe samples prepared in Example 1, mix the two probes in a ratio of 1:1, and shake well. Then react with wild-type (SEQ ID NO.3) and SNP site mutant (SEQ ID NO.4) target molecules respectively according to the molar ratio of 1:100, shake well, and incubate in an incubator at 25°C and 300rpm overnight.
[0069] Step 2: Place the mixture of the probe sample prepared in step 1, wild type (SEQ ID NO.3) and SNP site mutant (SEQ ID NO.4) in a temperature-controlled centrifuge (25°C, 12000r .p.m), remove the supernatant, precipitate out, redissolve with PBS buffer solution with a concentration of 0.1mol / L, and store in a refrigerator at 4°C for nanopore detection.
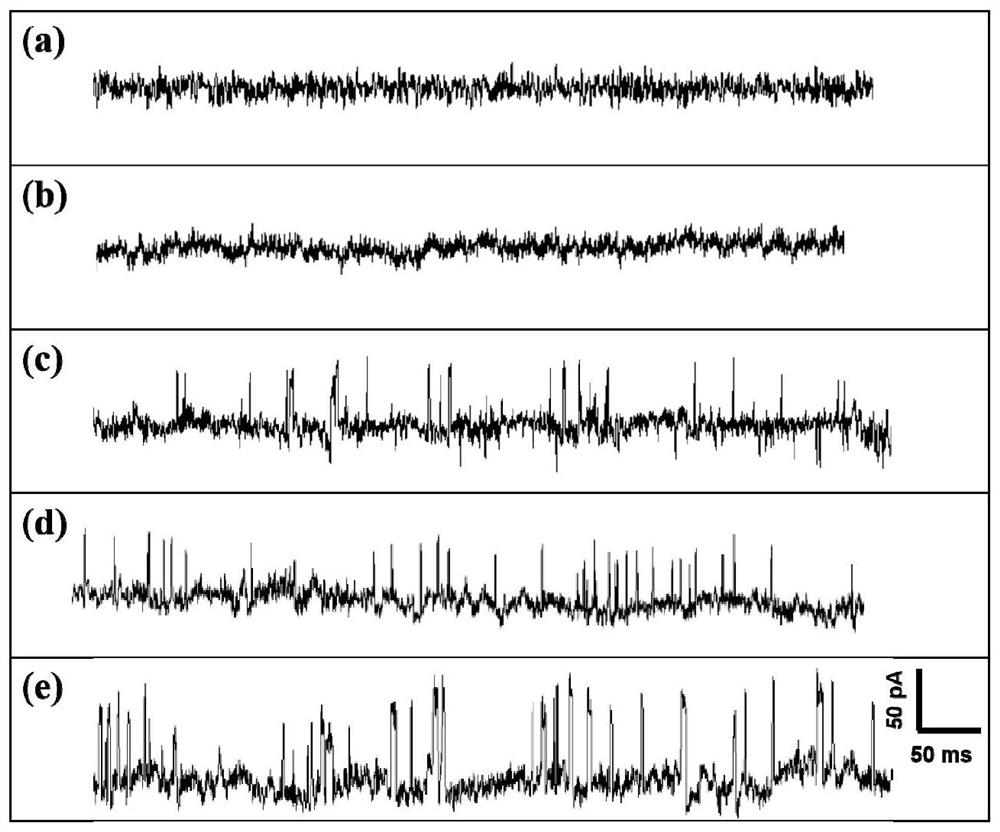
[0070] Step 3: Observing the complex of the prepared detection probe and the target molecule with a 40nm ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
| particle diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com