New application of drug screening cell model targeting HBV core promoter, Tricirine and structural analogue
A core promoter and cell model technology, applied in the fields of molecular biology and biomedicine, can solve problems such as drugs that cannot effectively inhibit the transcriptional activity of core promoters
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
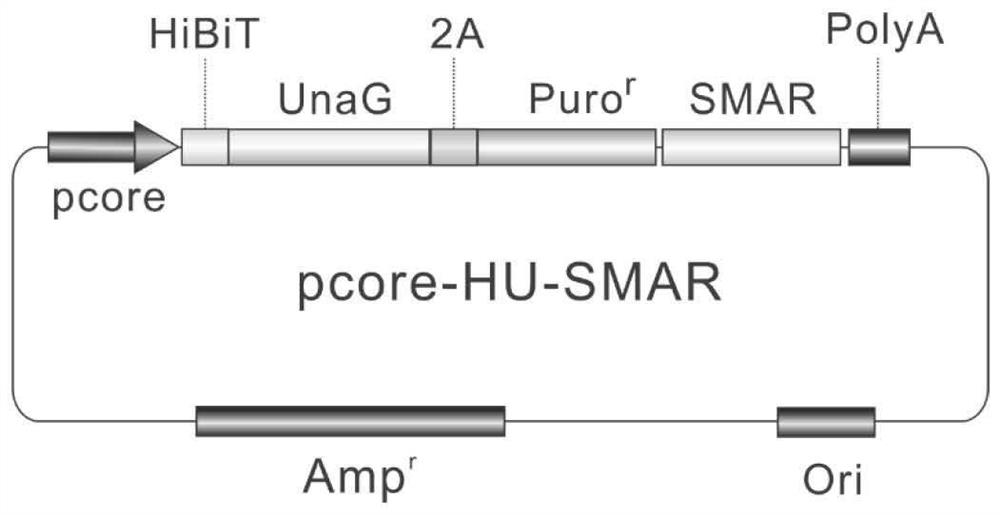
[0051] Example 1. Construction of a drug screening cell model targeting the HBV core promoter
[0052] 1. Design idea and working principle
[0053] The HBV core promoter functions on cccDNA, and cccDNA exists in cells in the form of minichromosomes independent of cell chromosomes. At this point, all cell models based on transgenes (integrated in the chromosomes of cells) are far from reality. To better model cccDNA transcription events, it is preferable to place the core promoter outside the chromosome.
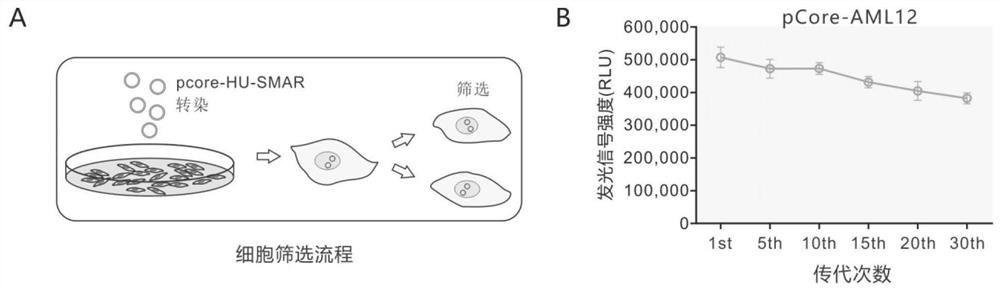
[0054] The cccDNA simulation system based on plasmid vector (or plasmid-like) transfection, such as the mcHBV-Gluc cccDNA model, and the integration-deficient lentiviral vector of Fluc based on Fluc, etc., are indeed located outside the chromosome, However, its significant defect is that the cccDNA mimic molecule cannot replicate, but is gradually lost with cell division and cannot exist stably. This results in the need for a transfection or infection step for each experi...
Embodiment 2
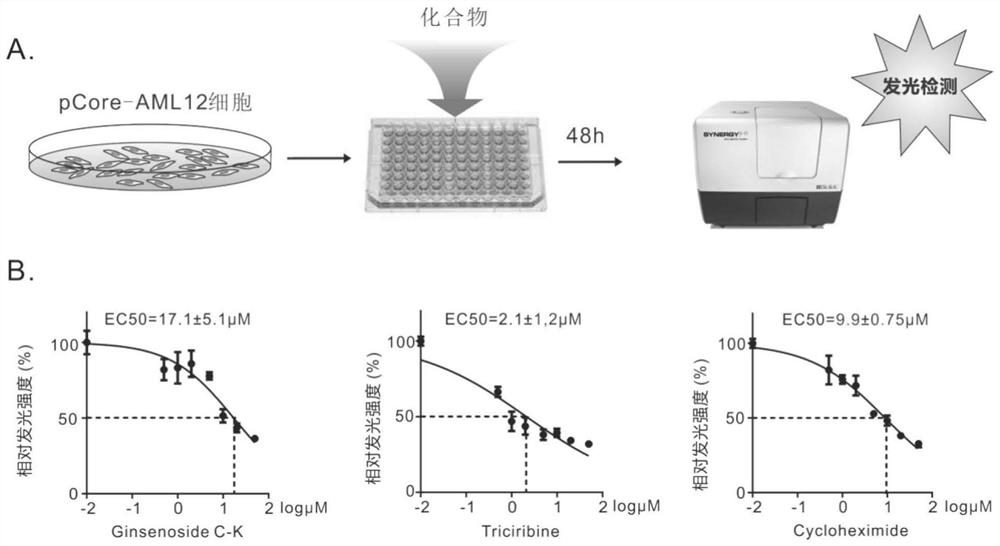
[0091] Example 2. Screening of compounds targeting HBV transcription
[0092] 1. Preliminary screening of compounds
[0093] After the stable passage cell pCore-AML12 was obtained, it was used for compound screening. The screening procedure is as follows: pCore-AML12 cells were revived and cultured in 10 cm culture dish with DMEM medium containing 10% fetal bovine serum (cultivated by conventional methods). After the cells have been passaged for 3 times, when the growth state is better, the cells are digested and seeded into 96-well plates. On the second day after cell inoculation, the compounds to be tested were respectively added to the 96-well plate with a final concentration of 5 μM, and two replicate wells were made for each compound. About 48 hours after adding the drug, aspirate the medium, add cell lysate to each well, incubate with shaking at room temperature for 20 minutes, then take the lysate and use HiBiT detection reagent for detection (the process is as follow...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com