Method for improving pig health by targeted inactivation of CD163
A technique for pairing, editing pigs, applied to other methods of inserting foreign genetic material, microbe-based methods, botanical equipment and methods, etc., can solve problems such as inability to be precise and efficient
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0217] This example illustrates the selection of target sites for knockout of the porcine CD163 gene in porcine cells.
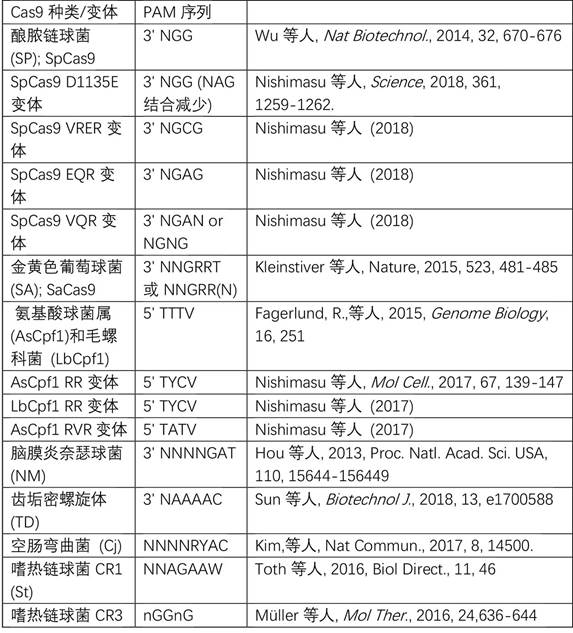
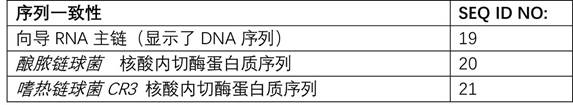
[0218]Single or paired delivery of guide RNAs by generating DNA sequence insertions, deletions, and combinations thereof in the porcine CD163 gene (Sscrofa11.1, GenBank Accession: GCA_000003025.6) using Streptococcus Cas9 / gRNA RNA-guided DNA endonuclease In combination with the protein, the sequence change in the CD163 gene reduces or abolishes the function, stability or expression of the CD163 messenger RNA or protein. The CD163 gene sequence from 120 nucleotides upstream of the translation initiation site (chr5:63300192) to 59 nucleotides downstream of CD163 exon 7 (chr5:63323390) was screened for genes with adjacent nGG or nGGnG Guide RNA binding sites for PAM sequences required for cleavage by the Cas9 protein from Streptococcus pyogenes or Streptococcus thermophilus CRISPR3 (Streptococcus thermophilus CR3), respectively, the sequences of which are prese...
Embodiment 2
[0220] This example demonstrates nucleofection to deliver a guide RNA / Cas9 endonuclease to porcine fetal fibroblasts.
[0221] To test the DNA cleavage activity of the edited porcine CD163 alleles generated in living cells, CRISPR-Cas endonucleases and guide RNAs targeting the sequences listed in Table 3 were nucleoffected into porcine fetal fibroblasts . To prepare porcine fetal fibroblast (PFF) cell lines from 28-35-day-old fetuses, mix 3.2 µg of Cas9 protein (S. The volume was 2.23 µl, which was then nucleoffected into PFF cells using a Lonza electroporator. In preparation for nucleofection, harvest PFF cells using trypsin expression (recombinant trypsin). After this, remove the medium from the cells, wash the medium once with HBSS or DPBS, and incubate at 38.5°C with trypsin for 3–5 min. Cells were then harvested with complete medium. Cells were pelleted by centrifugation (300 x g for five minutes at room temperature), the supernatant was discarded, and the cells were ...
Embodiment 3
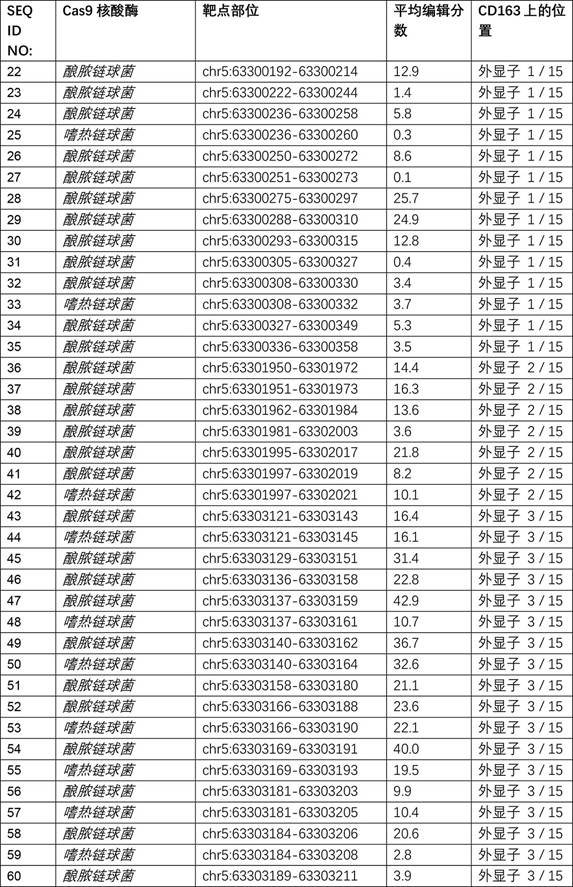
[0224] This example illustrates the editing frequency of a guide RNA / Cas9 combination directed to porcine CD163.
[0225] Nucleotide sequence changes were introduced into the porcine CD163 gene by delivering Cas9 protein complexed with guide RNAs to fetal fibroblasts, as described in Example 2.
[0226] To assess DNA double-strand cleavage at the porcine CD163 genomic target site mediated by the guide RNA / Cas endonuclease system, a region of genomic DNA approximately 250 bp around the target site was amplified by PCR and then passed through the amplicon Deep sequencing checks PCR products for edits. After 3 transfections, PFF genomic DNA was extracted as described in Example 2. The region surrounding the expected target site is PCR amplified with NEB Q5 polymerase and the sequence required for amplicon-specific barcoding is added, by two rounds of PCR, using tailed primers for ILLUMINA ® (ILLUMINA ® , San Diego, CA) sequenced. at ILLUMINA ® MISEQ ® Personal Sequencer (I...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com