Peptide fragment-based targeted proteome accurate quantification method
A proteomic and precise technology, applied in the field of targeted proteomics, can solve the problems of inconsistent antibody enrichment efficiency, high cost, and failure to screen out peptides, etc., to reduce time and economic costs, simple adjustment, and easy data processing Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
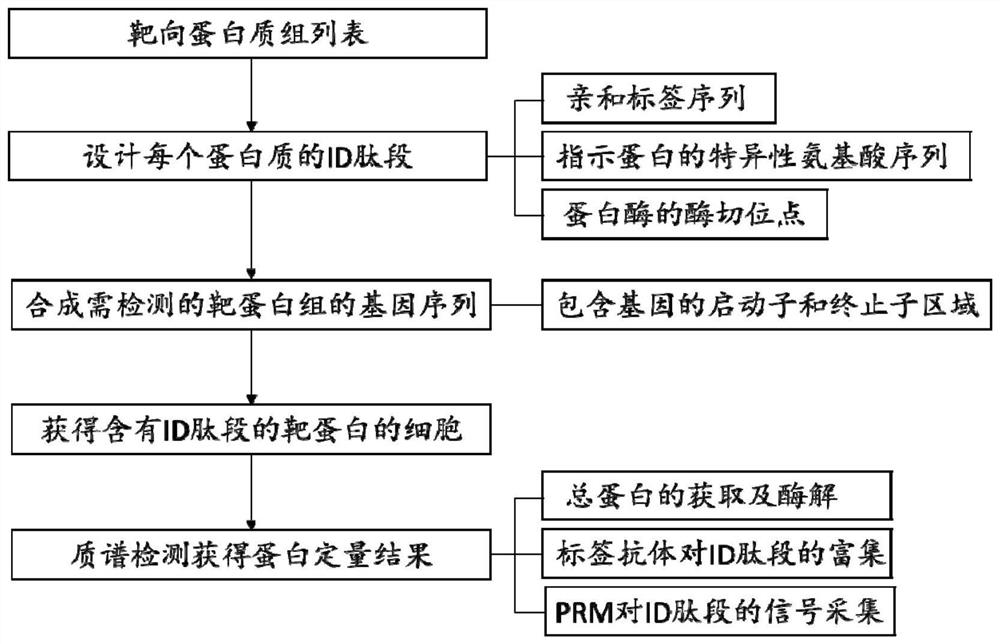
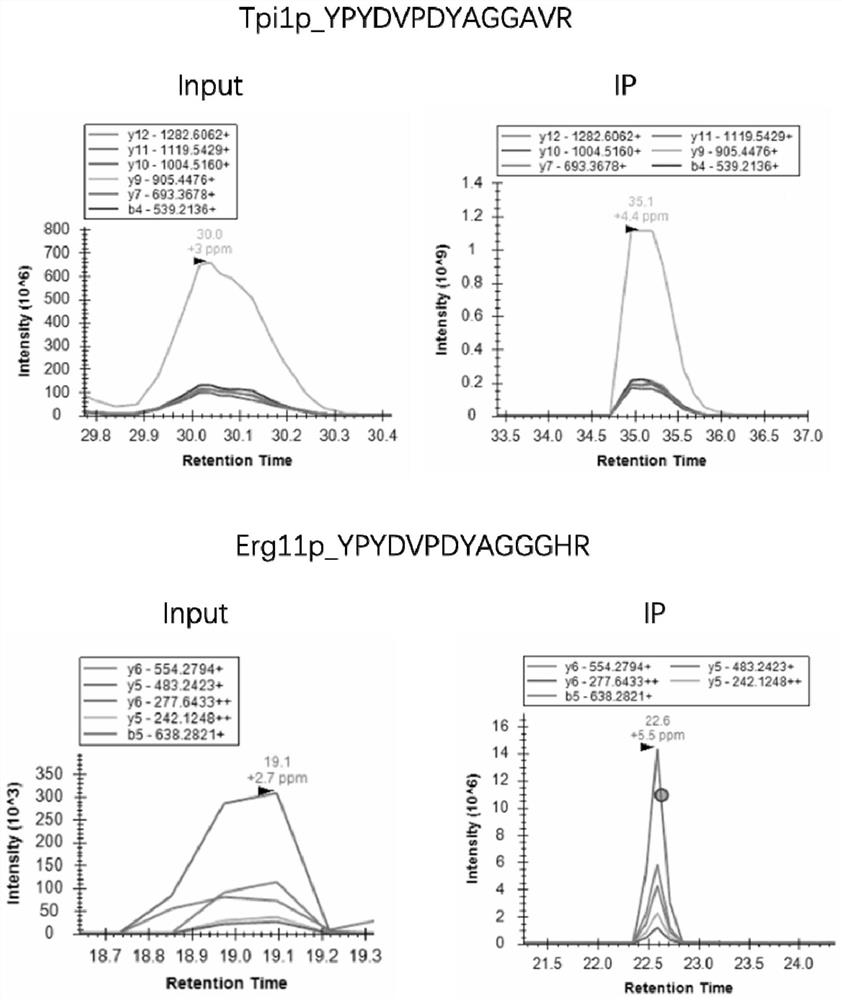
[0049] Example 1 selects 40 proteins in the yeast metabolic pathway, and uses the method of the present invention to detect the expression of these proteins.
[0050] (1) Design the ID peptide sequences corresponding to 40 proteins
[0051] The affinity tag sequence in the ID peptide is HA tag, the sequence is YPYDVPDYA, and the restriction site is arginine R, the trypsin restriction site. The design of the ID peptide is shown in Table 1:
[0052] Table 1 Amino acid sequences and DNA sequences of ID peptides corresponding to 40 proteins
[0053]
[0054]
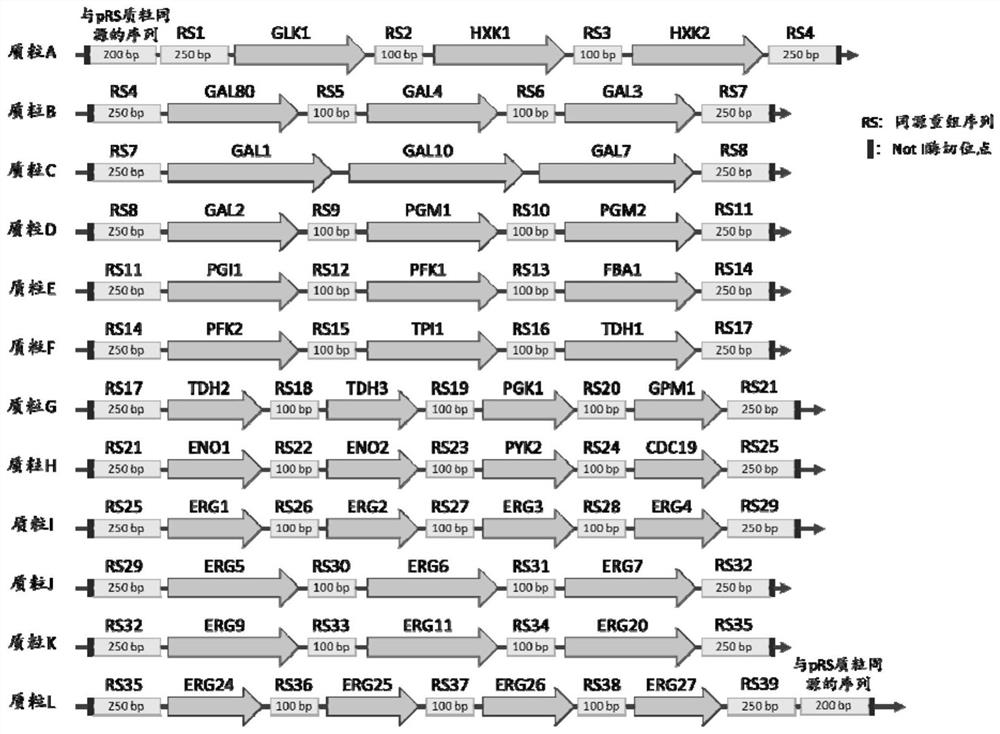
[0055] (2) Gene sequences of 40 proteomes to be detected for whole gene synthesis
[0056] The sequence of the ID peptide is added to the start codon of the corresponding gene ORF, and 3-4 genes are synthesized on a pMV plasmid through in vitro synthesis, and each gene contains a sequence of 500 bp upstream and 200 bp downstream of the ORF sequence. At the same time, 100bp random homologous recombination sequences w...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com