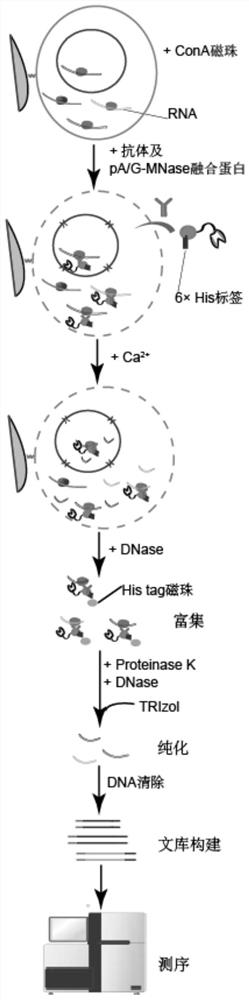
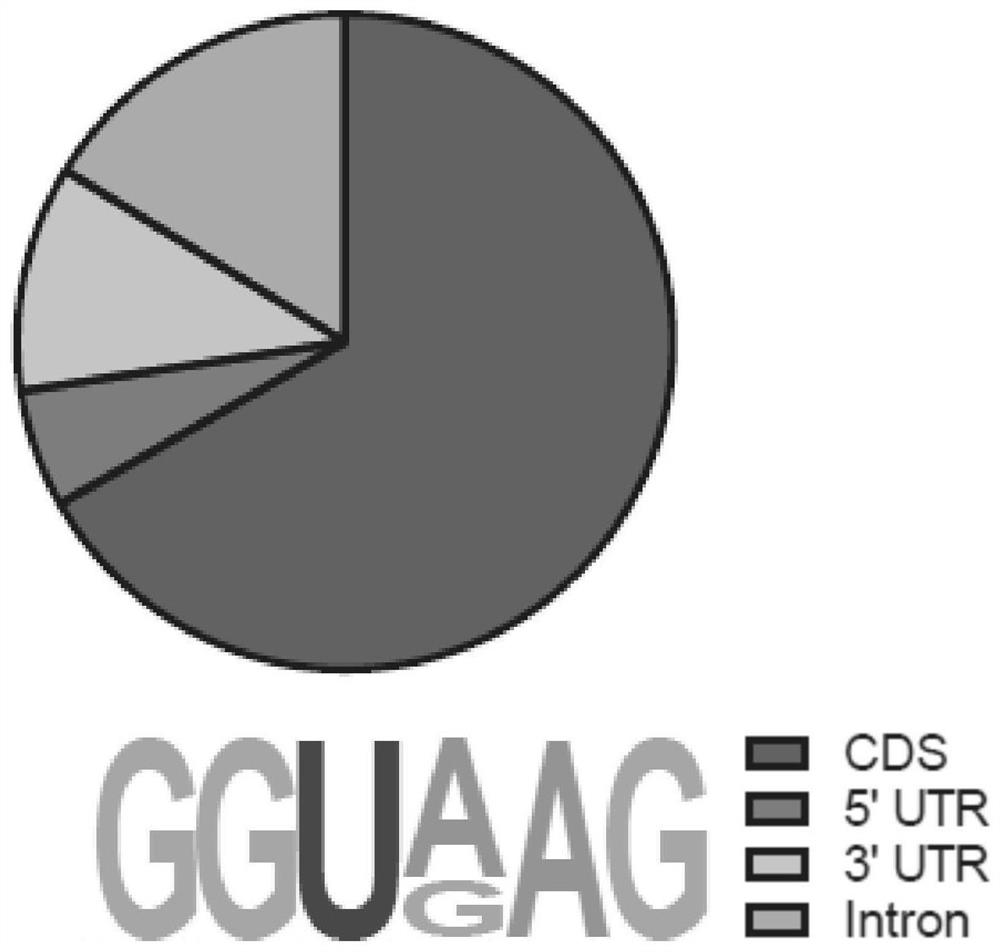
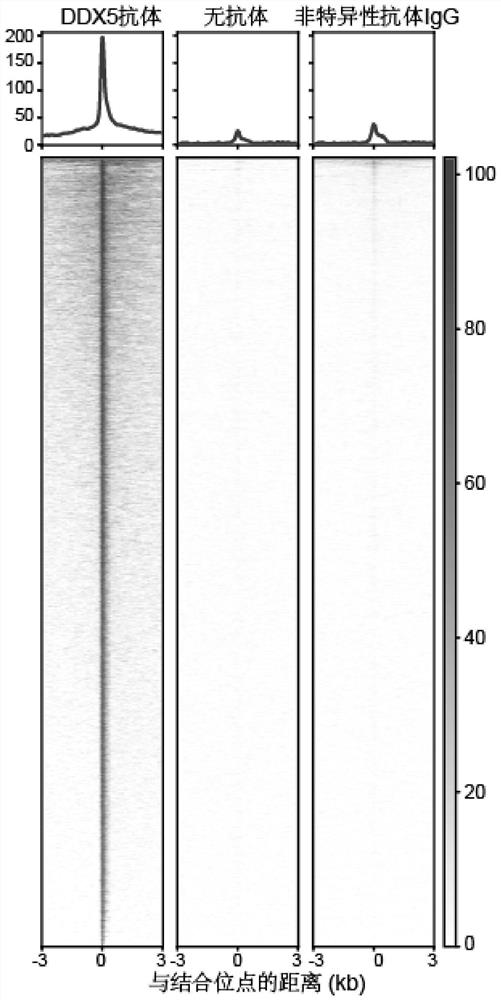
Method for obtaining target RNA of target RNA binding protein
A technology for binding proteins and fusion proteins, which is applied in the field of obtaining target RNA binding proteins and target RNAs, can solve the problems of non-in situ detection, large amount of cells, complicated steps, etc., to avoid the loss of many fragments, improve the quality of the library, and the method is fast simple effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0089] (1) Collect 100,000 mouse embryonic stem cells cultured in vitro, wash them once with PBS (PH=7.2), centrifuge at 600g room temperature for 3min, remove the supernatant, and wash once with 200μL wash buffer;
[0090] (2) 8 μL Con A beads (Bangslabs) were washed with binding buffer and mixed with the cells in (1) and incubated for 10 min, centrifuged at 100 g at 4°C, discarded the supernatant (magnetic stand), and washed with wash buffer. Add 1 μg DDX5 protein antibody to 100 μL washbuffer, 0.5 μL 5% Digitonin (digitonin), 1 μL protease inhibitor (100×), incubate at 4°C for 2-12 hours;
[0091] (3) Add 0.07 μg / μL fusion protein and incubate at 4°C for 2 hours;
[0092] The DNA coding sequence of the 6×His tag is placed at the end of the DNA coding sequence of the fusion protein precursor through plasmid construction. Introduce the expression plasmid into expression strains such as BL21 or DE3, and purify the fusion protein to obtain the tagged protein. The fusion prote...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com