Primer for monitoring DNA of fish environment at middle and upper reaches of Yarlung Zangbo River
A fish and upstream technology, applied in the field of molecular ecology, can solve problems such as the inability to monitor the level of fish species in the middle and upper reaches of the Brahmaputra, and achieve the effects of easy operation, accurate and comprehensive detection, and simple steps.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Acquisition of primers for environmental DNA monitoring of fish in the middle and upper reaches of the Yarlung Zangbo River:
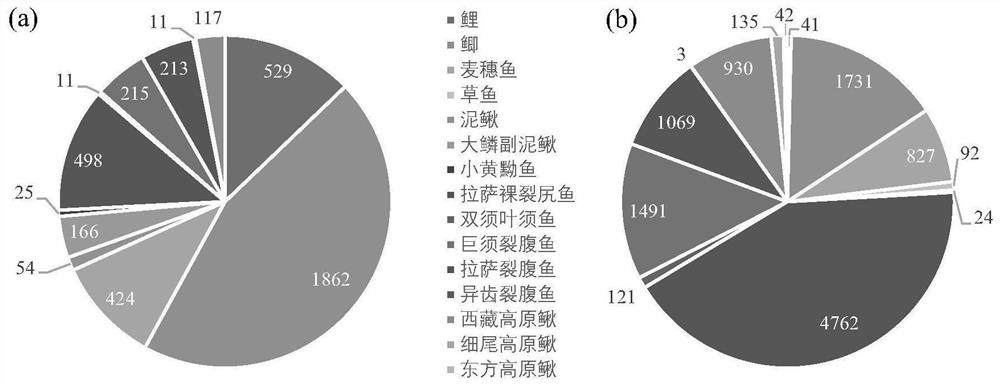
[0033] The applicant collected the fish list in the middle and upper reaches of the Yarlung Zangbo River through field investigation and literature review, including 15 indigenous species and 14 exotic species (Table 1), belonging to 4 orders and 10 families. Species of different purposes, but also closely related species of the same genus.
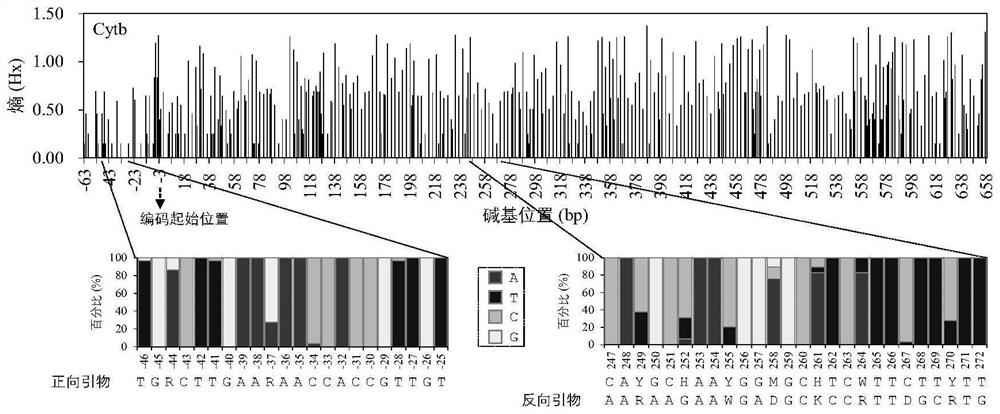
[0034] The primers used in environmental DNA technology need to be relatively conservative, so as to ensure effective amplification in all species; and the amplicon sequence needs to be diverse, so as to ensure the identification effect on all species. The mitochondrial 12S, COI and Cytb gene sequences of all fish in the middle and upper reaches of the Yarlung Zangbo River were collected by DNA sequencing and NCBI database. The average K2P genetic distances within species were 0.06%, 0.20% and 0.34%, respec...
Embodiment 2
[0040] Application of primers in monitoring fish species diversity in the middle and upper reaches of the Yarlung Zangbo River:
[0041] (1) 1 L of water was collected in the Lalu Wetland in Lhasa (sample point A) and the Yarlung Zangbo River in Zha Nang County (sample point B), and the DNA was collected on a mixed cellulose filter membrane by a vacuum filter pump, and Store cryopreserved in liquid nitrogen.
[0042] (2) Extract the water sample eDNA using the Powerful Water Sample DNA Extraction Kit, and the water sample DNA concentrations in sample points A and B are 10.5ng / ul and 6.6ng / ul, respectively.
[0043] (3) Using water sample eDNA as a template, edm_YZR_Cytb-F and edm_YZR_Cytb-R as primers, and using NEB Q5 ultra-fidelity DNA polymerase for the first PCR amplification, the reaction system is: 12.5ul Q5 ultra-fidelity 2× Master Mix, each 1.0ul forward and reverse primers, 5.0ul DNA template, 5.5ul ddH 2 O. The reaction parameters were: pre-denaturation at 95°C fo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com