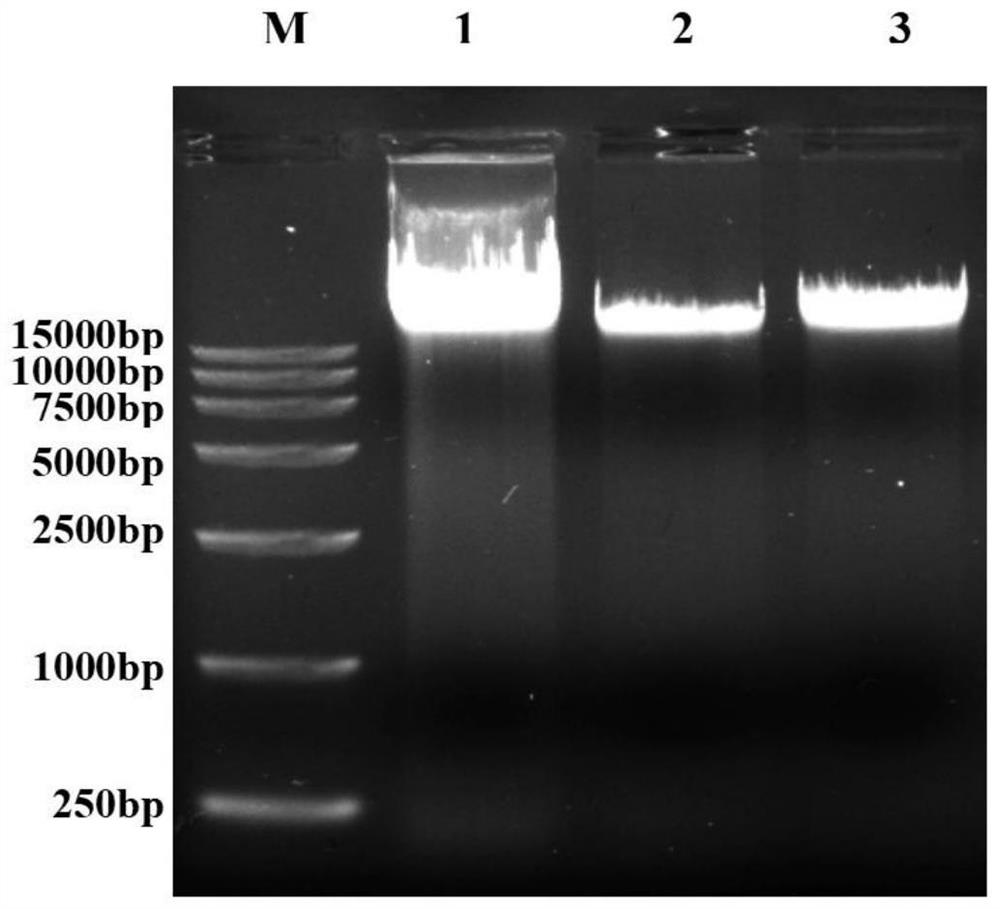
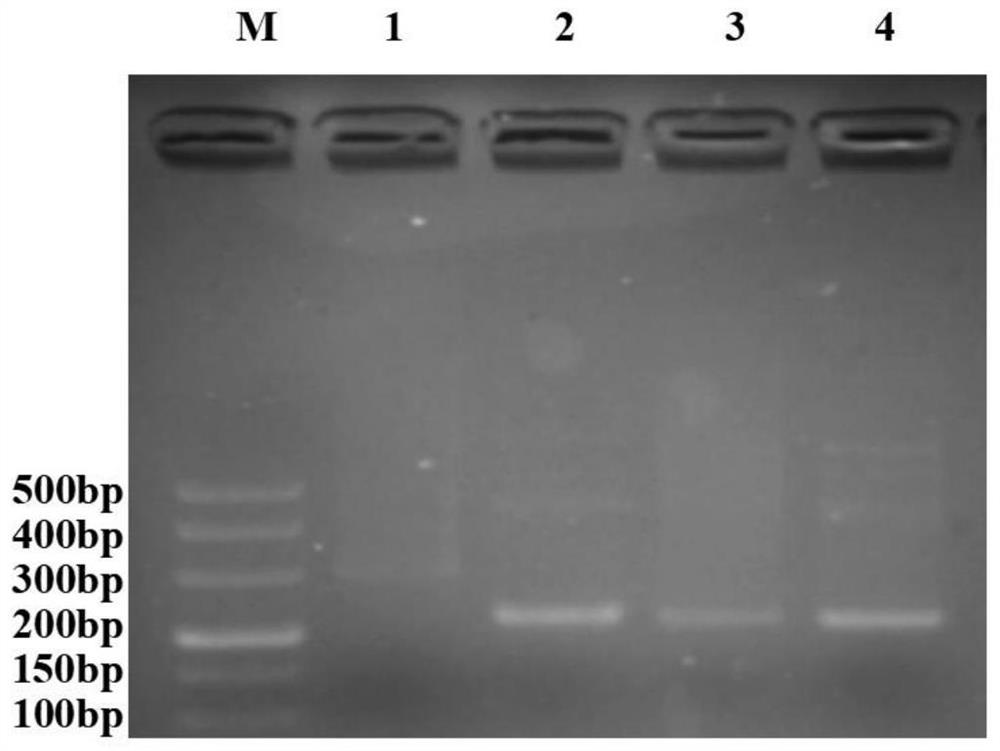
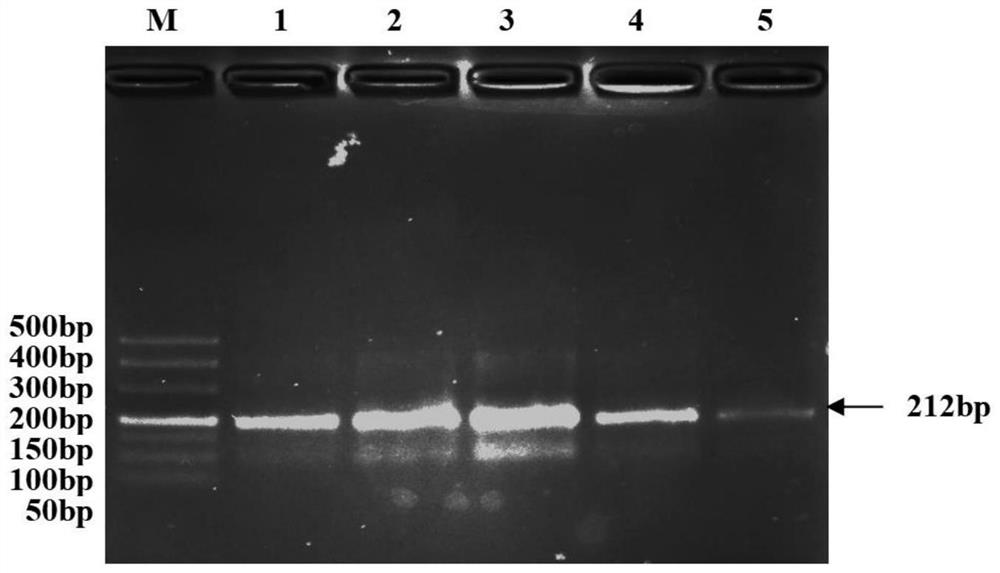
Method for identifying transgenic Bt gene based on recombinase polymerase isothermal amplification method
A technology of recombinant enzyme polymerase and constant temperature amplification method, which is applied in the direction of biochemical equipment and methods, microbial measurement/inspection, etc. It can solve the problems that PCR technology cannot implement rapid on-site detection, high operating technical requirements, and high instrument costs. , to achieve the effect of being conducive to on-site detection, low technical requirements, and good specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] (1) Extraction of corn genomic DNA:
[0034] 1) Take 0.4 g of corn leaves from which the stems have been removed, put them into a pre-cooled mortar with liquid nitrogen added (the amount of liquid nitrogen added accounts for 2 / 3 of the volume of the mortar) and grind, pass through a 80-mesh sieve, Load into a 5mL centrifuge tube before the temperature rises;
[0035] 2) Add the improved 2×CTAB extract solution (0.1g / 100mL Tris-HCL, 1mL / 100mL β-mercaptoethanol, 3g / 100mL CTAB, 60mmol / L EDTA, 1.2mol / L NaCl) preheated at 65°C and add 0.04g PVP ) 1.6mL, incubate at 65°C, shake once every 5min, for a total of 30min;
[0036] 3) After the incubation, take it out, set the centrifuge at 4°C, 12000rpm, put the centrifuge tube into it carefully to avoid DNA damage, and centrifuge for 5min;
[0037] 4) Aspirate 1.4 mL of the supernatant with a micropipette, add a mixture of chloroform and isoamyl alcohol equal to the volume of the supernatant (wherein the volume of chloroform and...
Embodiment 2
[0052] (1) Extraction of corn genomic DNA:
[0053] 1) Take 0.4 g of corn leaves from which the stems have been removed, put them into a pre-cooled mortar with liquid nitrogen added (the amount of liquid nitrogen added accounts for 2 / 3 of the volume of the mortar) and grind, pass through a 90-mesh sieve, Load into a 5mL centrifuge tube before the temperature rises;
[0054] 2) Add the improved 2×CTAB extract solution (0.1g / 100mL Tris-HCL, 1mL / 100mL β-mercaptoethanol, 3g / 100mL CTAB, 60mmol / L EDTA, 1.2mol / L NaCl) preheated at 65°C and add 0.04g PVP ) 1.6mL, incubate at 65°C, shake once every 5min, for a total of 30min;
[0055] 3) Take it out after the incubation, set the centrifuge at 4°C, 12000rpm, put the centrifuge tube into it carefully to avoid DNA damage, and centrifuge for 8 minutes;
[0056] 4) Aspirate 1.4 mL of the supernatant with a micropipette, add a mixture of chloroform and isoamyl alcohol equal to the volume of the supernatant (wherein the volume of chloroform...
Embodiment 3
[0071] (1) Extraction of corn genomic DNA:
[0072] 1) Take 0.4 g of corn leaves from which the stems have been removed, put them into a pre-cooled mortar with liquid nitrogen added (the amount of liquid nitrogen added accounts for 2 / 3 of the volume of the mortar) and grind, pass through a 100-mesh sieve, Load into a 5mL centrifuge tube before the temperature rises;
[0073] 2) Add the improved 2×CTAB extract solution (0.1g / 100mL Tris-HCL, 1mL / 100mL β-mercaptoethanol, 3g / 100mL CTAB, 60mmol / L EDTA, 1.2mol / L NaCl) preheated at 65°C and add 0.04g PVP ) 1.6mL, incubate at 65°C, shake once every 5min, for a total of 30min;
[0074] 3) Take it out after the incubation, set the centrifuge at 4°C, 12000rpm, put the centrifuge tube into it carefully to avoid DNA damage, and centrifuge for 10min;
[0075] 4) Aspirate 1.4 mL of the supernatant with a micropipette, add a mixture of chloroform and isoamyl alcohol equal to the volume of the supernatant (wherein the volume of chloroform an...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com