Application of phage depolymerase ORF38 protein in pasteurella multocida capsule typing identification
A Pasteurella, typing and identification technology, applied in the biological field, can solve problems such as Pasteurella that have not yet been found
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
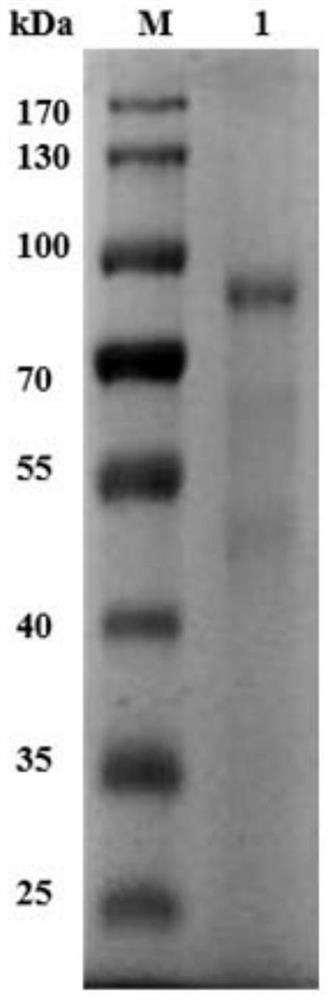
[0028] The construction of embodiment 1 recombinant protein
[0029] 1. Gene Amplification
[0030] The target fragment gene ORF38 (Gene ID: 54980989) was amplified using phage PHB01 as a template.
[0031] Forward primer: 5'-CCG GAATTC ATGTCATCAAGATATCAGTA-3'EcoRI
[0032] Reverse primer: 5'-GC GTC GAC TCACAATTGACACCATTTATTCA-3'SalI
[0033] The amplification system and amplification conditions are as follows:
[0034]
[0035] PCR reaction program: pre-denaturation at 98°C for 5 min; pre-denaturation at 98°C for 20 s, annealing at 60°C for 20 s, extension at 72°C for 2 min, 30 cycles; final extension at 72°C for 10 min, and storage at 4°C.
[0036] 2. Construction of recombinant plasmids
[0037] (1) 200ng of the recovered target fragment and 100ng of the recovered pET-28a vector were digested with restriction endonucleases EcoRI and SalI at 37°C for 2h;
[0038] (2) Ligate the digested product overnight at 4°C under the action of T4 ligase;
[0039] (3) After...
Embodiment 2
[0059] Activity and stability of embodiment 2 protein
[0060] 1. Dot-spot assay for protein activity
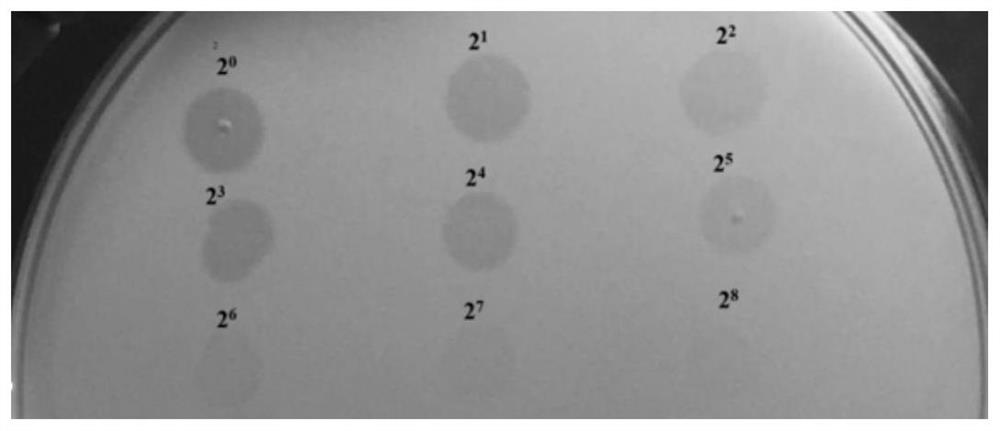
[0061] Take 300μL PmD bacterial solution and mix it with 6mL TSB medium containing 0.75% agar, pour the lower plate, and after the plate is naturally dried, dilute the purified depolymerase ORF38 protein by two times, and take 5 μL (0.15 μg) of ORF38 protein was spotted onto the prepared double-layer plate, and incubated at 37°C for 12 hours to observe the results.
[0062] The result is as figure 2 As shown, the cleavage activity can still be seen when the protein is diluted to the 8th power.
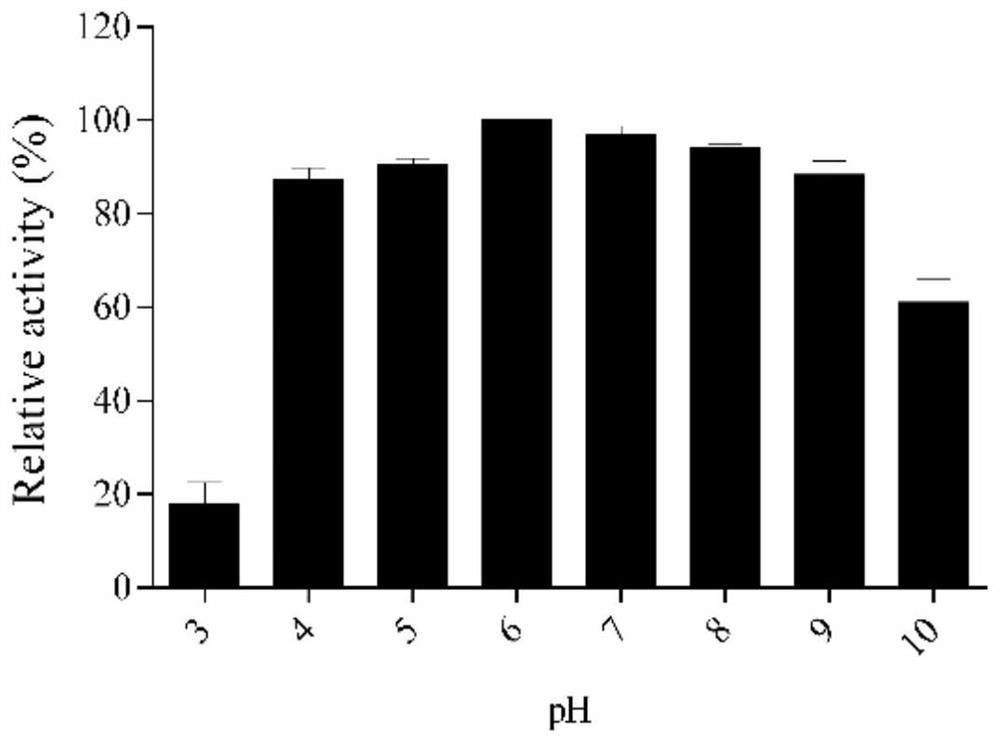
[0063] 2. Protein pH stability
[0064] PmD exopolysaccharide (EPS) was extracted by hot phenol method. The lyophilized EPS was dissolved by adding appropriate amount of PBS buffer solution with different pH to reach the required concentration. Take 900 μL EPS diluent (EPS is 5 mg / mL) and mix with 100 μL ORF38 protein (30 μg / mL), and incubate at 37° C. for 1 h. The degradation...
Embodiment 3
[0069] Capsule typing of embodiment 3 Pasteurella multocida
[0070] Of the 55 strains of PmD preserved in our laboratory, 10 strains of Pm with capsular serology A, 5 strains of Pm with capsular serology B, 1 strain of Pm with capsular serology E, and 1 strain of Pm with capsular serology For Pm of F, 10 strains of Bordetella bronchis, 10 strains of Salmonella, 10 strains of Escherichia coli, 10 strains of Staphylococcus aureus, 10 strains of Haemophilus parasuis and 10 strains of Streptococcus were cultured to the logarithmic phase, and 300 μL of the above After the bacterial solution was mixed with 6 mL of TSB medium containing 0.75% agar, the lower plate was poured. After the plate was dried, the ORF38 protein was diluted with PBS (pH=6.0) buffer solution, 5 μL (0.15 μg) of ORF38 protein was spotted onto the double-layer plate, and incubated upside down in a 34°C-38°C incubator for 12 hours.
[0071] Table 1 is the ORF38 protein cleavage profile test. Using 5 μL (0.15 μg...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com