Genome selection method for breeding Plectropomus leopardus disease-resistant improved variety
A technology of genome selection and eastern star spot, applied in biochemical equipment and methods, recombinant DNA technology, microbial measurement/inspection, etc., can solve the problems of missing pedigree information, low selection accuracy, lack of family materials, etc., to improve survival rate Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] Example 1: Establishment of Eastern star spot disease-resistant reference population
[0024] Juveniles of East star spot were collected from a number of large-scale breeding farms in Hainan and Shandong, and the collected juveniles were transported to places where artificial infection experiments could be carried out, and these juveniles were treated by intraperitoneal injection of Vibrio harveyi bacteria liquid artificial infection. Consider the time of injection of the bacterial solution as the starting point (0 o'clock), observe the survival of the juvenile fish after injection every 8 hours, remove the dead individual in time, record the individual information (such as full length, body weight, and death time, etc.) and cut off the tail fin Fin rays were stored in absolute ethanol. The experiment was carried out for 5 days. After the end, the caudal fin rays of surviving individuals were collected and stored in absolute ethanol, and the data of individual origin, ...
Embodiment 2
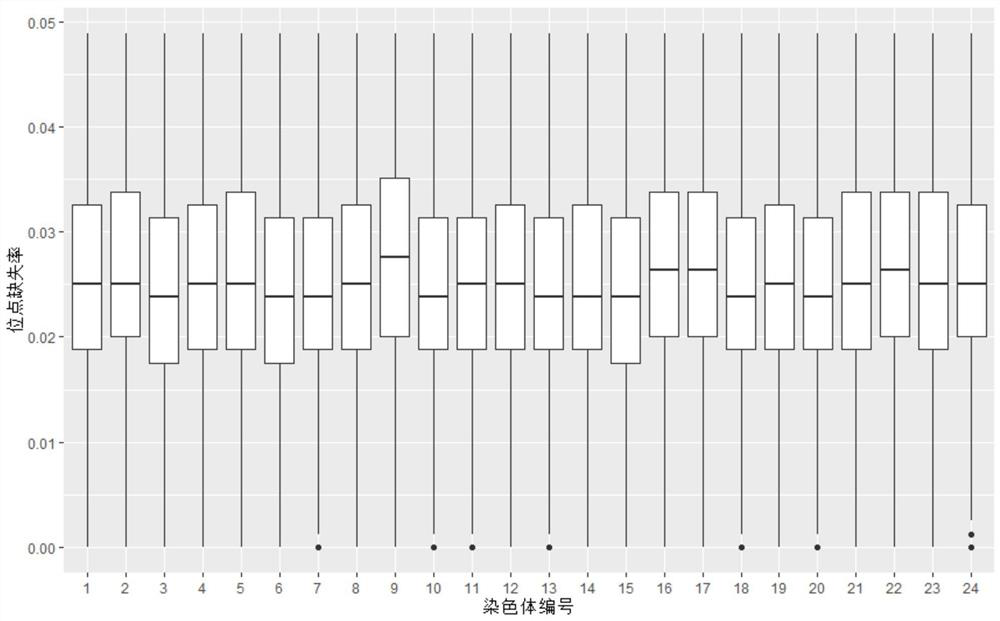
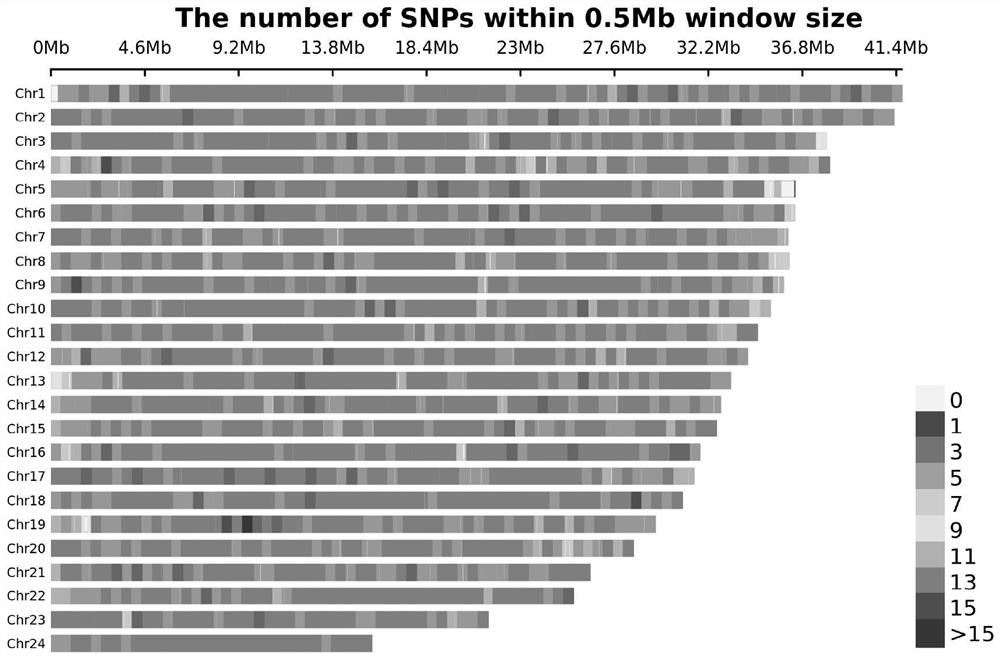
[0028] Example 2: Whole-genome resequencing and screening of high-quality SNPs in the eastern star spot disease-resistant reference population
[0029] 1. Reference population whole genome resequencing and mutation detection
[0030] Genomic DNA from the reference population of Eastern star spot disease resistance was extracted and purified using the standard procedure provided by the DNA extraction kit (TIANGEN, Beijing). In addition to the genomic DNA of the reference population, the genomic DNA of 298 Eastern star spots with a disease-resistant phenotype was also extracted to verify the effect of the method of the present invention. Purified genomic DNA was used to build an Illumina paired-end library, and then the Illumina HiSeq 2000 sequencing platform was used for sequencing. After filtering out low-quality sequencing reads (reads), the software BWA was used to compare these reads to the Eastern Star Spot reference genome. Use the software GATK to detect the variation i...
Embodiment 3
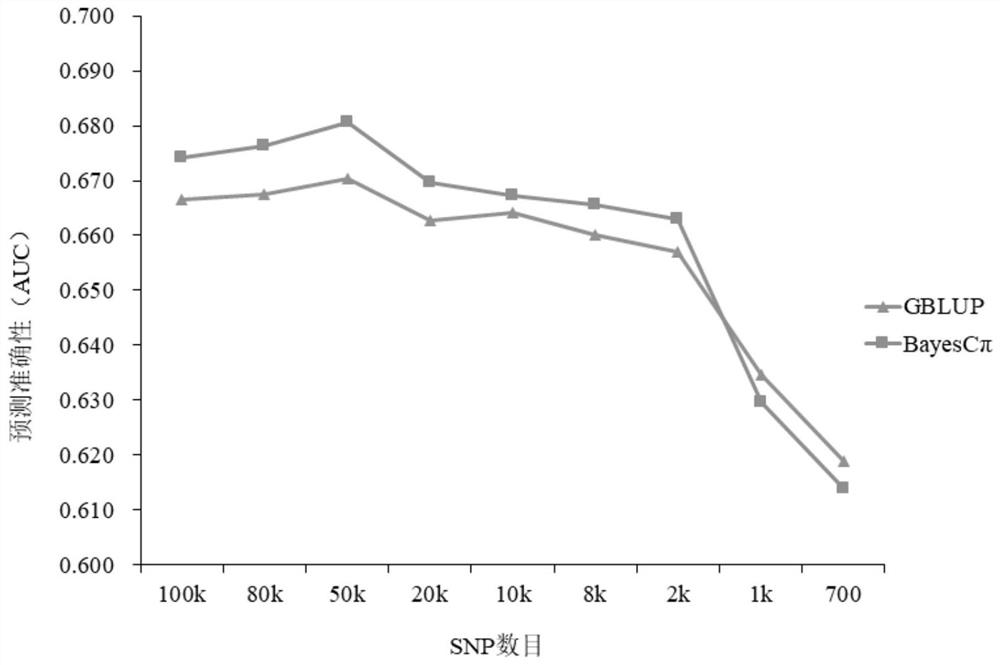
[0039] Example 3: Accuracy of two gene-wide selection methods in predicting disease-resistant GEBV in eastern star spot
[0040] In order to explore the method and the appropriate number of SNP markers for estimating the GEBV resistance of Eastern star spot against Harvey vibrio disease, the present invention compared the G- Trends of BLUP and Bayes Cπ in predicting the accuracy of GEBV resistance to harvei vibrio disease in eastern star spot as the number of SNPs decreases. The area under the receiver operating characteristic curve (AUC) was used as the evaluation index for the prediction accuracy, and the calculation was carried out 25 times, and the final accuracy was expressed as the mean of 25 times. The following R packages need to be installed before calculation: data.table, ASReml, BGLR, Rcpp, pROC and parallel.
[0041] 1. Extract different numbers of SNP collections (SNP subsets) from the eastern star spot disease-resistant reference population
[0042] Different num...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com