SSR marker for detecting bruchid-resistant variety of broad beans and application of SSR marker
A technology for resisting weevil and broad bean, which is applied to the determination/inspection of microbes, DNA/RNA fragments, biochemical equipment and methods, etc., and can solve the problems of low attention, unsequenced genome sequence, and difficulty in resisting weevil varieties, etc. problems, to achieve significant selectivity, low equipment requirements, and great application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Embodiment 1-acquisition of SSR core motif
[0027] Improving the method for extracting broad bean genome DNA, comprising the steps of:
[0028] (1) Take 80 mg of fresh young leaf tissue of broad bean, add 700 μl of DNA extraction solution and 1% PVP, grind thoroughly, and transfer the grinding solution into a corresponding 1.5 mL centrifuge tube; polyvinyl pyrrolidone (PVP for short);
[0029] (2) Place the centrifuge tube in a 65°C water bath for 30 minutes, take it out every 5 minutes and shake gently to mix;
[0030] (3) After the water bath is completed, add 500 μl of phenol / chloroform / isoamyl alcohol (v / v / v is 25:24:1) into the fume hood, mix it upside down, centrifuge at 12000rpm for 10min, and transfer the supernatant to a new centrifuge tube, then add an equal amount of chloroform / isoamyl alcohol (v / v is 24:1), mix up and down;
[0031] (4) Centrifuge at 12000rpm for 10min, take 500μl of the supernatant and transfer it to a new centrifuge tube, add 2 times th...
Embodiment 2
[0040] Embodiment 2-SSR core motif nucleotide information
[0041] Using PacBio third-generation full-length sequencing technology combined with RNA-seq method, the whole genome of broad bean was sequenced, and the sequence of SEQ ID NO.7 was copied to MISA online software (http: / / pgrc.ipk-gatersleben.de / misa / ) , using the default parameters for analysis, it was found that the 48bp-67bp nucleotide sequence at the 5' end contains the SSR core motif of (TC)10, and the nucleotide sequence is shown in SEQ ID NO.7:
[0042] 5’-GTTATTGTCGTGGAGAAAGGAAAACCTCTCCCGCCACGAAAATTCTATTCTCTCTCTCTCTCTCTCTCTGTAGTAGCCATATACACAGCTAGCTTCGCGTCACAATCTCACTCTCATCAACTCTTCAAACTAATGGCTGCAACTCTTCAAACTCTGCGACTTCCTTTTCATCCATCTATCACACCCCTAAATTCATATCCTTTCCCAACCTCTACCGTCCATTACTCCCCCAAATCAAGCACCTTCAAGGGTTCATCTGTCGCCACCCGCAACAAACCAGTACCTTCTTCCAAAGCTTCCAATTCGCAGTACAGTCCCACTGTTACCGAAAATCTCGCTGACATTAGCATTTTCTCGGCCGCCGGTGAGCCCGTCATGTTCAAAGATCTTTGGGACCAAGAACAGGAAAAGCCCTGATCTTTATCCATTTCTTATCTGTTTTGGGCACAACCATTTTGACCTAA...
Embodiment 3
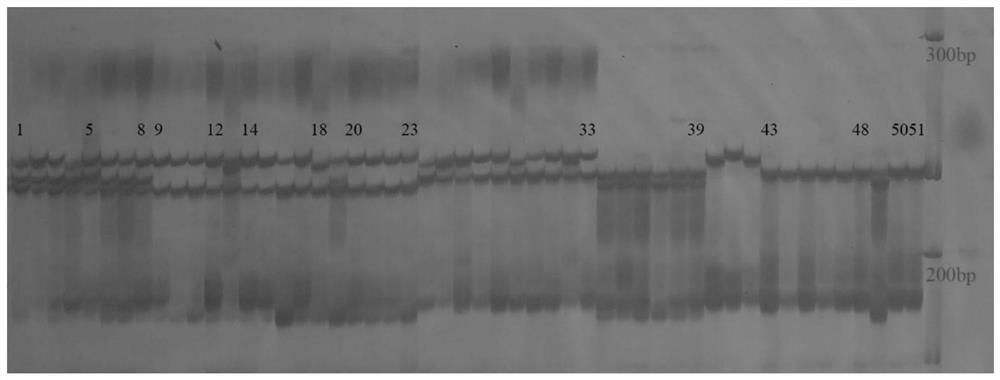
[0043] Example 3 - Using SSR markers to assist in the identification of broad bean varieties resistant to bean weevil
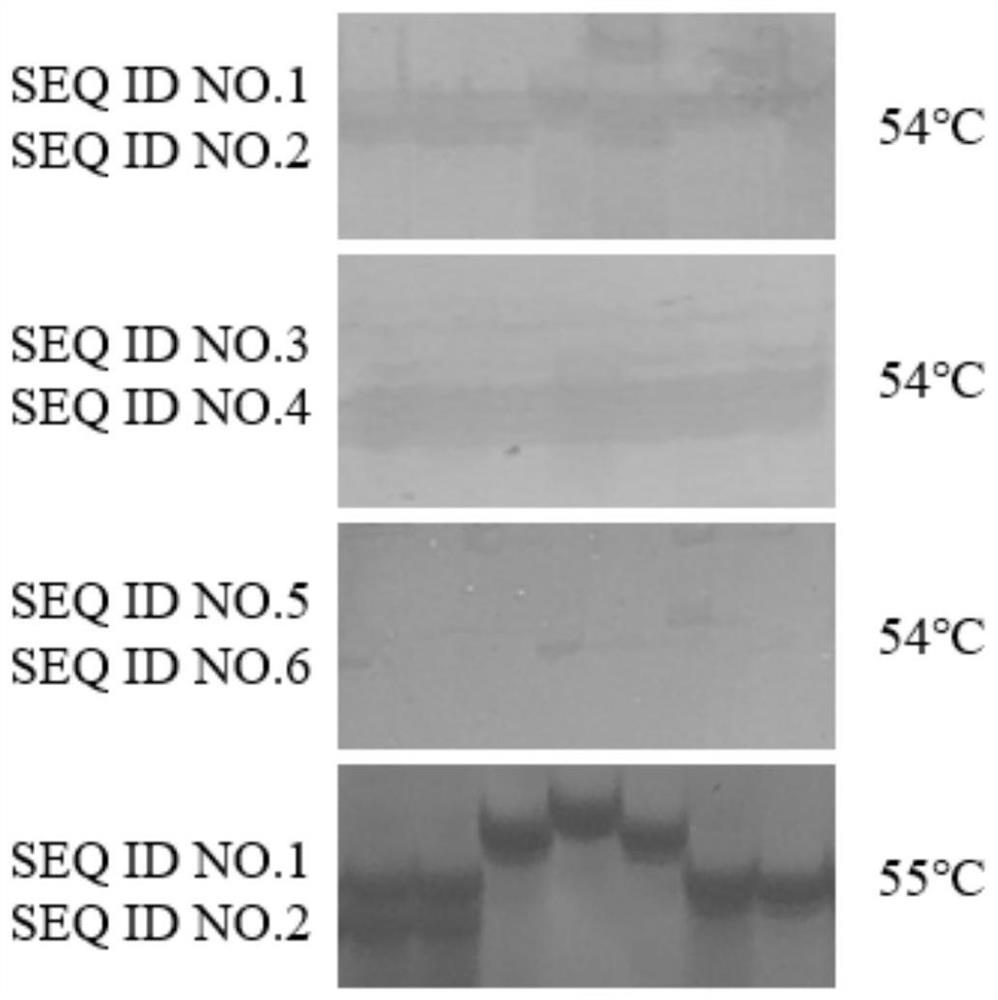
[0044] Step 1, primer design: According to the nucleotide sequence SEQ ID NO.7 of the specific faba bean genome containing SSR core motif (TC) 10, 3 pairs of SSR primers were designed with Primer 3 software, the sequence is:
[0045] SEQ ID NO.1: AGAAAGGAAAACCTCCCG;
[0046] SEQ ID NO. 2: GTGCTTGATTTGGGGGAGTA. (first pair of primers)
[0047] SEQ ID NO.3: AGAAAGGAAAACCTCCCG;
[0048] SEQ ID NO. 4: GAGTGAGATTGTGACGCGAA. (Second pair of primers)
[0049] SEQ ID NO.5: AGAAAGGAAAACCTCCCG;
[0050] SEQ ID NO. 6: AAGGTACTGGTTTGTTGCGG. (third pair of primers)
[0051] Step 2, PCR amplification: PCR conditions were explored.
[0052] The initial annealing temperature of the primer was obtained by lowering the Tm value given in the primer synthesis sheet by 3°C. Using part of the broad bean genomic DNA to be tested as a template, the selected DNA template was...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com