Method for increasing mass spectrum identification number of protein and/or peptide fragment group by using polystyrene material
A polystyrene-based, identification method technology, applied in the field of proteomics, to achieve the effects of reducing human error, simple operation, and avoiding interference
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] 1. Take out different plasma samples from the refrigerator, take 200 μL of plasma from each sample and thaw at 37°C, centrifuge at 1,500g for 10 minutes at 4°C, remove the bottom precipitate and transfer the supernatant to a new tube;
[0034] 2. Take 100 μL plasma and add 100 μL binding buffer (50 mM Tris, 10 mM EDTA) to resuspend polystyrene microspheres (particle size 1 μm, CAS number: 9003-53-6), and incubate at room temperature for 10-60 minutes;
[0035] 3. After the incubation is completed, centrifuge at 12,000g for 10 minutes to remove the supernatant, resuspend the pellet with 300 μL washing buffer reagent (50mM Tris, 10mM EDTA), incubate at room temperature for 5 minutes, and centrifuge at 12,000g for 5 minutes to remove the supernatant;
[0036] 4. Repeat step 3 twice;
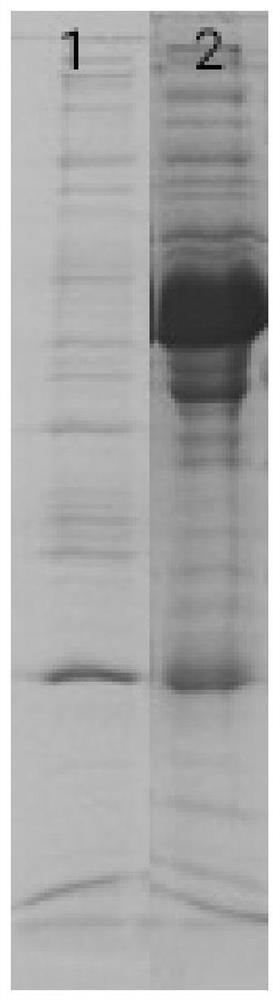
[0037] 5. Add the precipitate obtained by combining the original plasma and materials into protein electrophoresis Loading Buffer and boil, then run electrophoresis on the supernatant, the re...
Embodiment 2
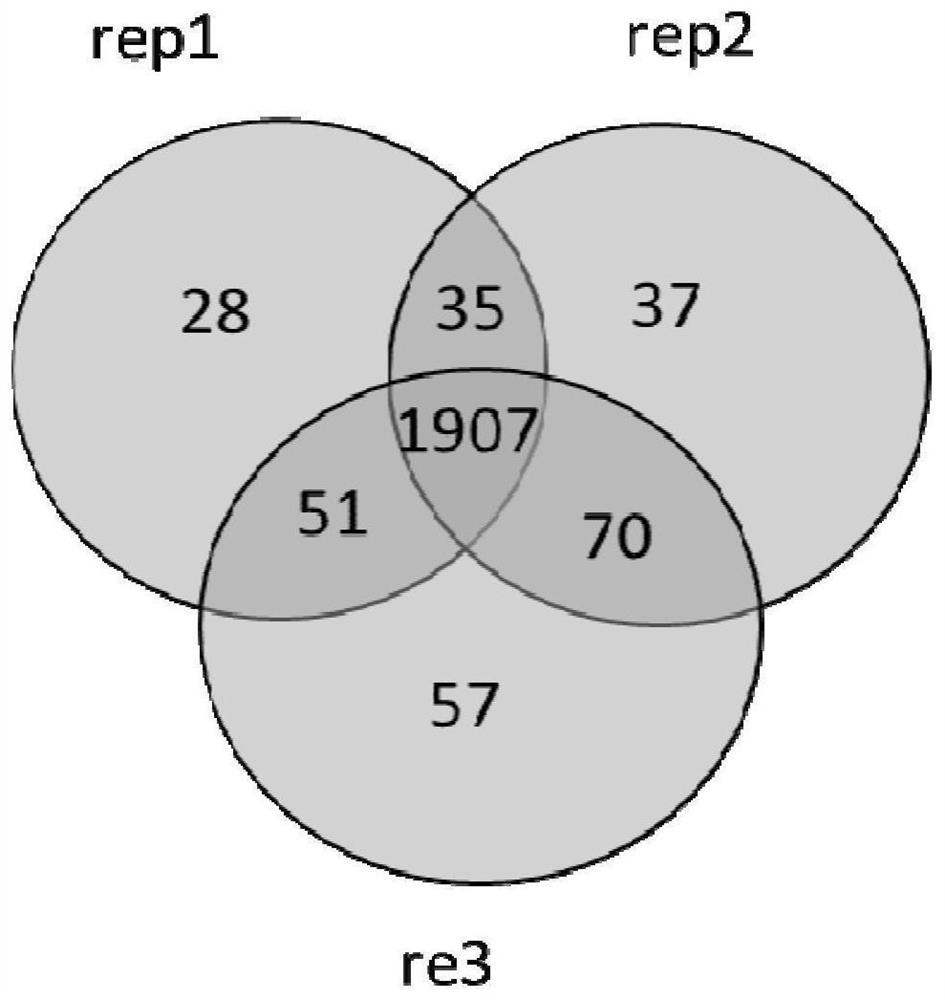
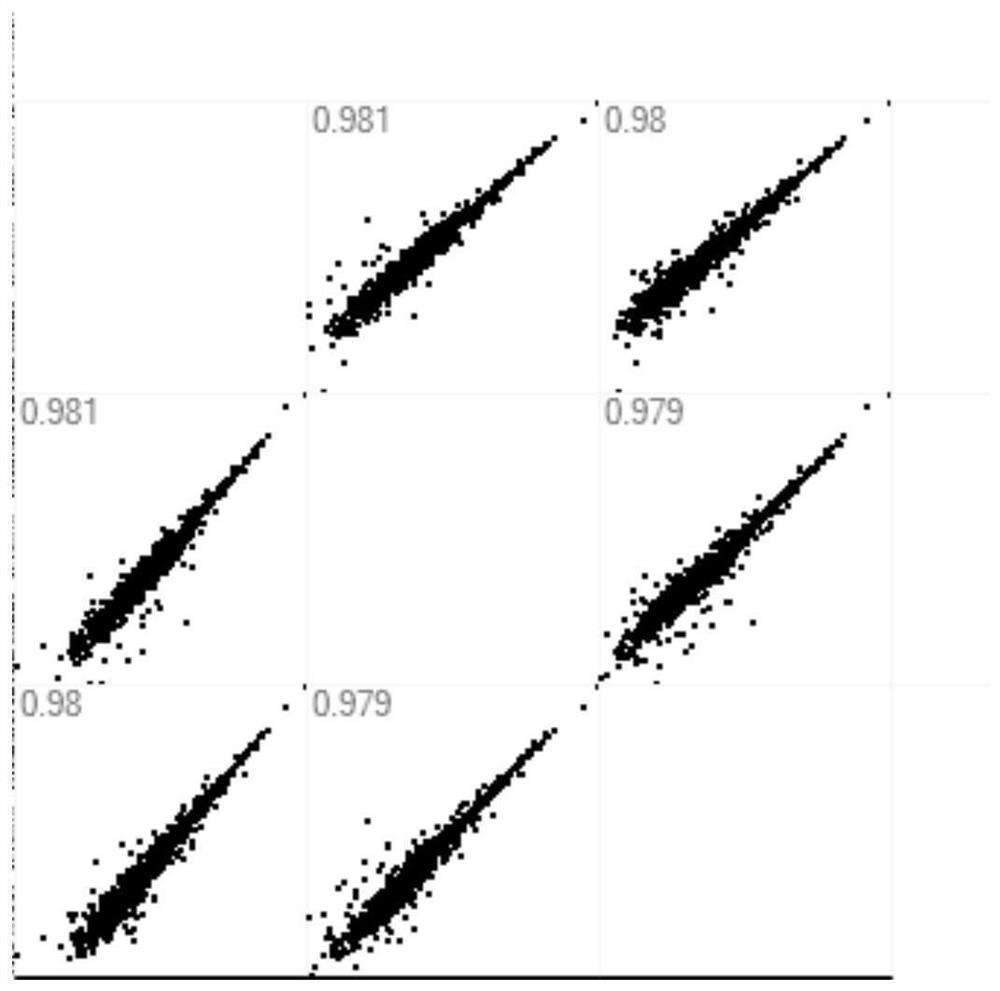
[0060] In order to prove that the use of this material can be used for the enrichment of micro samples and enzymatic desalination at the protein level or peptide level, it can be used to replace the existing traditional SP3 mixed magnetic bead materials. In this example, the SP3 mixed material was used as the control group, and the polystyrene material in Example 1 was used as the carrier to conduct proteomic analysis experiments on Hela cell protein and peptide extracts:
[0061] Take the Hela cell extract protein standard as an example:
[0062] 1. Take two 100 μg Hela cell protein samples dissolved in 40uL buffer (4% SDS, 100mM Tris, pH 8.0) for reductive alkylation;
[0063] 2. Add 10μL stored 10μg / μL SP3 mixed beads and the same amount of polystyrene microspheres (particle size between 50nm and 100μm, CAS number: 9003-53-6), then add 150uL buffer and 200uL acetonitrile As a binding buffer, shake and mix gently, and react at room temperature for 8 minutes;
[0064] 3. Af...
PUM
| Property | Measurement | Unit |
|---|---|---|
| particle diameter | aaaaa | aaaaa |
| particle diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com