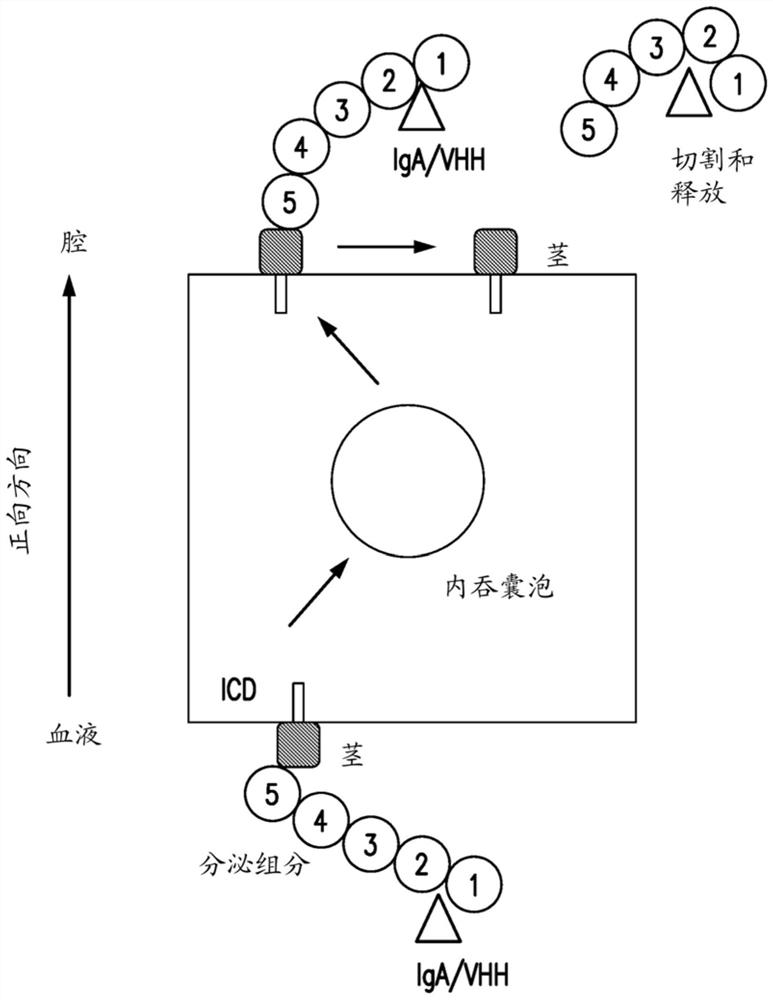
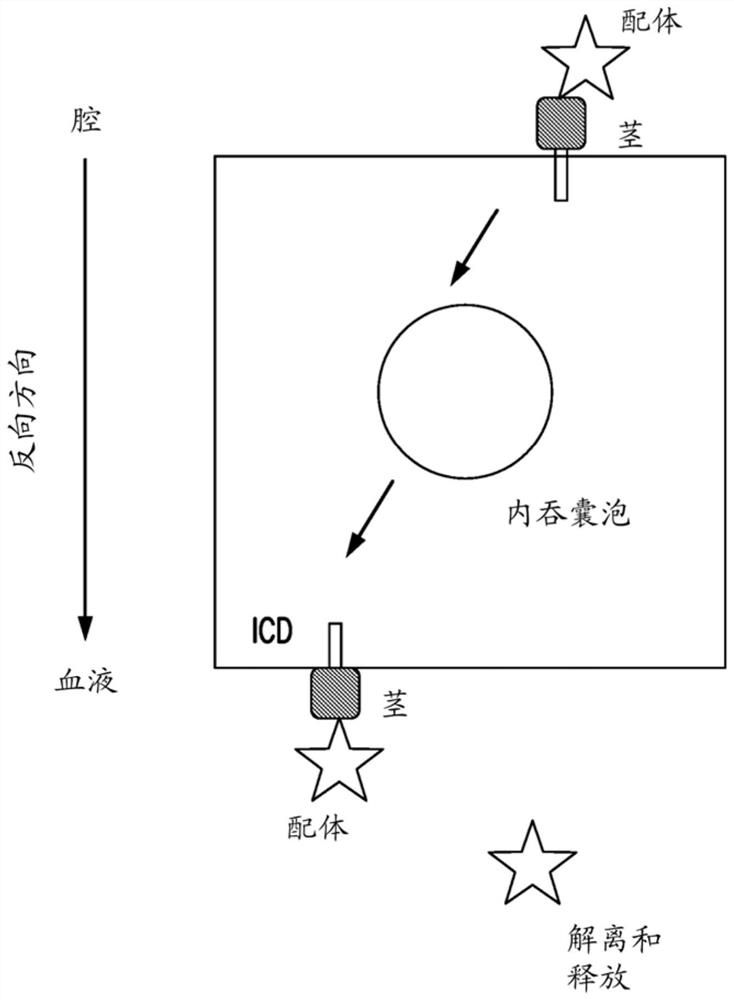
Materials and methods for multidirectional biological transport
A single-domain antibody, molecular technology, applied in the field of materials and methods for multidirectional biological transport, which can solve problems such as restricted transport
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0568] 5.2.6. Preparation of single domain antibody
[0569] Single domain antibodies provided herein can be produced by culturing cells transformed or transfected with a vector containing a nucleic acid encoding a single domain antibody. Polynucleotide sequences encoding the polypeptide components of the antibodies of the present disclosure can be obtained using standard recombinant techniques. Desired polynucleotide sequences can be isolated and sequenced from antibody producing cells such as hybridoma cells or B cells. Alternatively, polynucleotides can be synthesized using nucleotide synthesizers or PCR techniques. Once obtained, the sequence encoding the polypeptide is inserted into a recombinant vector capable of replicating and expressing the heterologous polynucleotide in a host cell. Many vectors available and known in the art can be used for the purposes of this disclosure. Selection of an appropriate vector will depend primarily on the size of the nucleic acid ...
Embodiment 1
[0700] 6.1. Example 1: Immunization, Recovery and Screening of pIgR Conjugates
[0701] To generate a panel of single domain antibodies that bind to pIgR, llamas were immunized with recombinant human pIgR (hpIgR) and / or mouse pIgR (mpIgR) for approximately 90 days. Whole blood and PBMC were isolated from llamas and RNA was prepared. After first-strand eDNA synthesis, llama-specific primers annealing to (i) VH (heavy chain variable region), (ii) VHH leader sequence gene, and (iii) CH2 gene were used to PCR amplify the VH and VHH gene pools.
[0702] The VHH pool was separated from the VH pool by running the PCR fragments on a gel and excising the smaller bands. The VHH gene pool was reamplified and cloned into CMV-based mammalian vectors. VHH-genes are formatted as Ig-fusions. The library was transformed into E. coli. Single colonies were picked in a 96-well format for Sanger sequencing. From approximately 300 unique sequences, a selected number of VHH sequences were sel...
Embodiment 2
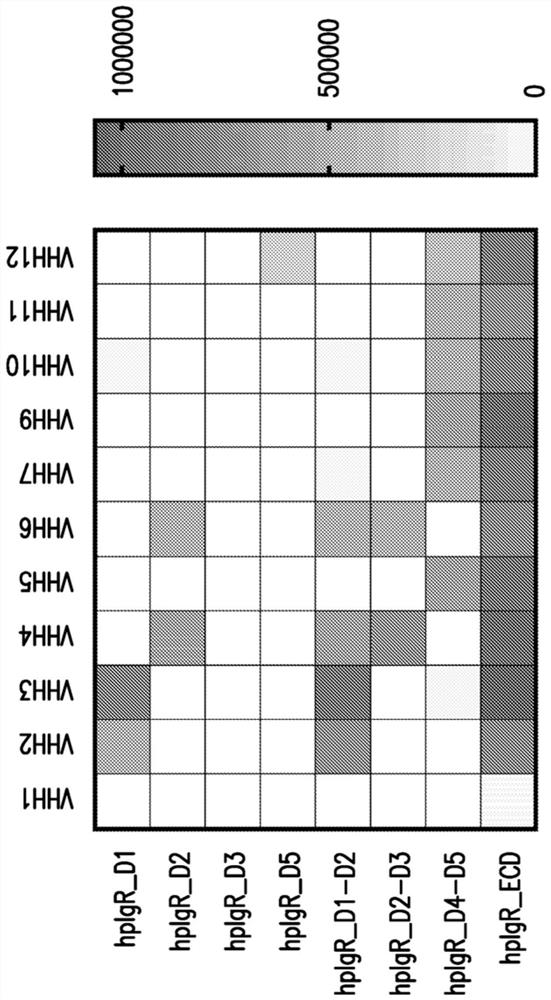
[0713] 6.2. Example 2: Biophysical characterization of hpIgR-specific binders
[0714] Ten plgR binders from Example 1 (8 hpIgR specific and 2 human / mouse cross-reactive) were selected for further biophysical and functional assays. Ten pIgR binders were expressed and purified from CHO cells using protein-A affinity chromatography. Size-exclusion chromatography combined with multi-angle light scattering revealed 10 VHH-mono-Fc conjugate species (VHH2, VHH3, VHH4, VHH5, VHH6, VHH7, VHH9, VHH10, VHH11, and VHH12) in the molecular weight range of 41.3 kDa to 48.7kDa.
[0715] The thermal stability of the samples was determined by differential scanning fluorimetry, specifically the NanoDSF method, using an automated Prometheus instrument. Measurements were performed by loading samples from a 384-well sample plate into a 24-well capillary. Duplicate runs were performed for each sample. The Prometheus NanoDSF user interface (Fusion Scan tab) was used to set the experimental par...
PUM
| Property | Measurement | Unit |
|---|---|---|
| melting point | aaaaa | aaaaa |
| melting point | aaaaa | aaaaa |
| melting point | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com