Difunctional marking method of 4-sulfydryl uracil and application of difunctional marking method in sequence enrichment and single base resolution sequencing
A technology of mercaptouracil and labeling method, applied in biochemical equipment and methods, microbial determination/inspection, organic chemistry, etc., can solve problems such as non-enrichment, and achieve the effect of high reaction efficiency and good biocompatibility
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
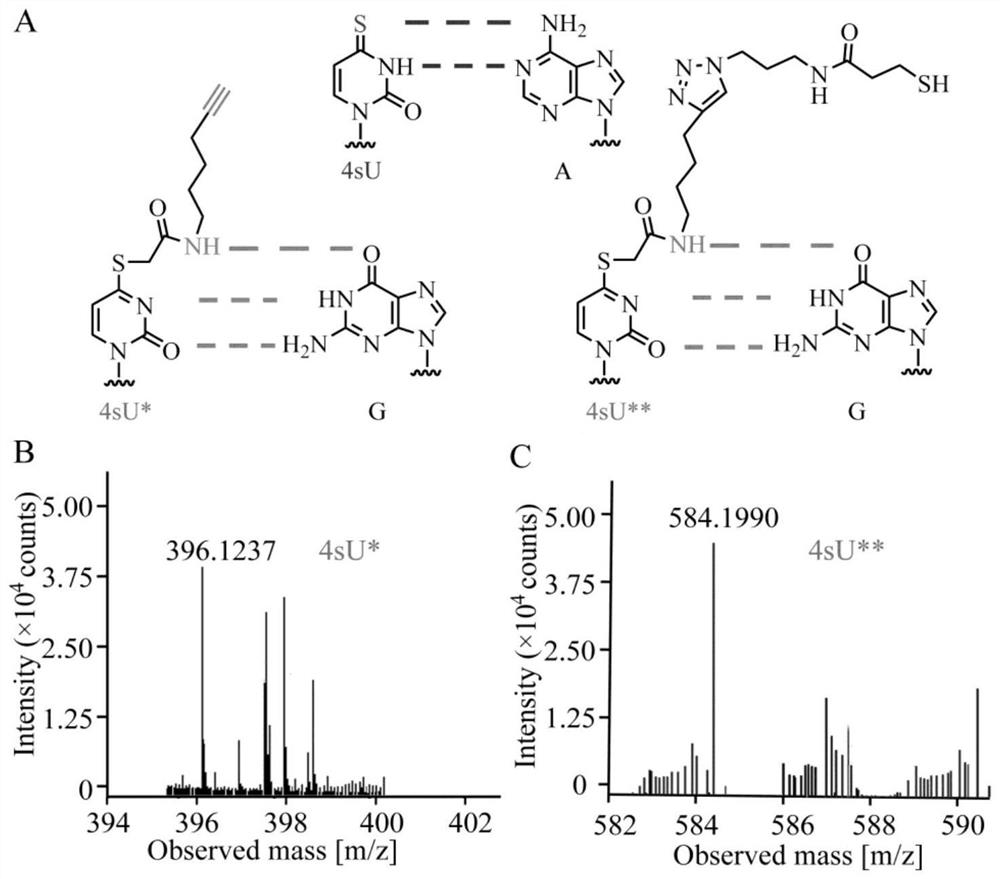
[0036] Using the nucleophilic substitution reaction of E-IAA to 4-mercaptouracil, the dual-functional labeling of 4-mercaptouracil and its nucleic acid samples can be realized, which can not only realize the enrichment of target sequences but also perform single-base resolution on them.
[0037] In vitro transcription of RNA with 4-mercaptouracil, the specific process is: sequence cDNA Sigle A or cDNASigle G or cDNA mult A or cDNA mult G and cDNA T7 for DNA extension, followed by transcription under the action of T7 RNA polymerase containing 4- The RNA of mercaptouracil, the nucleophilic substitution reaction of RNA and E-IAA. The reaction conditions are 10% DMSO aqueous solution as a solvent, sodium phosphate buffer with a pH value equal to 8, and reaction at 50° C. for 30 min. After the reaction, the RNA was recovered by alcohol precipitation. The labeled RNA was reacted with rSAP and RNAse I at 37°C for 12 hours to degrade the nucleic acid into nucleosides. Finally, vario...
Embodiment 2
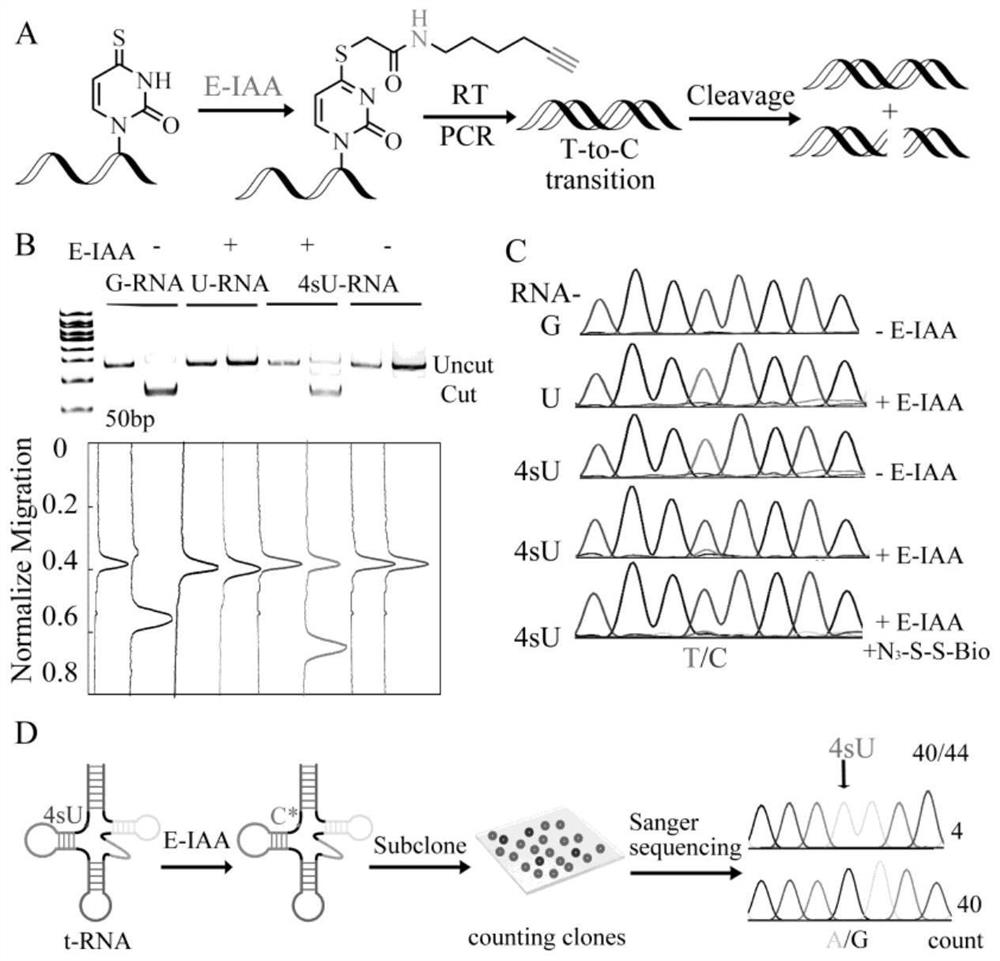
[0039] The nucleophilic substitution reaction of 4-mercaptouracil by using E-IAA described in the present invention is used to perform single base resolution on the nucleic acid sequence of 4-mercaptouracil.
[0040] The RNA with 4-mercaptouracil was transcribed in vitro, and the RNA was reacted with E-IAA. The reaction conditions were 10% DMSO aqueous solution as solvent, sodium phosphate buffer with a pH value equal to 8, and reacted at 50° C. for 30 minutes. After the reaction, the RNA was recovered by alcohol precipitation. The primers used for reverse transcription of labeled RNA are RT primer, and the primers used for PCR are PCRfp1, PCR rp1, PCR fp2 and PCR rp2. PCR products were characterized by 15% PAGE and sequencing. A mutation from T to C has occurred at the original position of 4-mercaptouracil. Therefore, this method realizes the single base recognition of 4-mercaptouracil. Such as figure 2 As shown, 15% PAGE and one-generation sequencing characterize the sin...
Embodiment 3
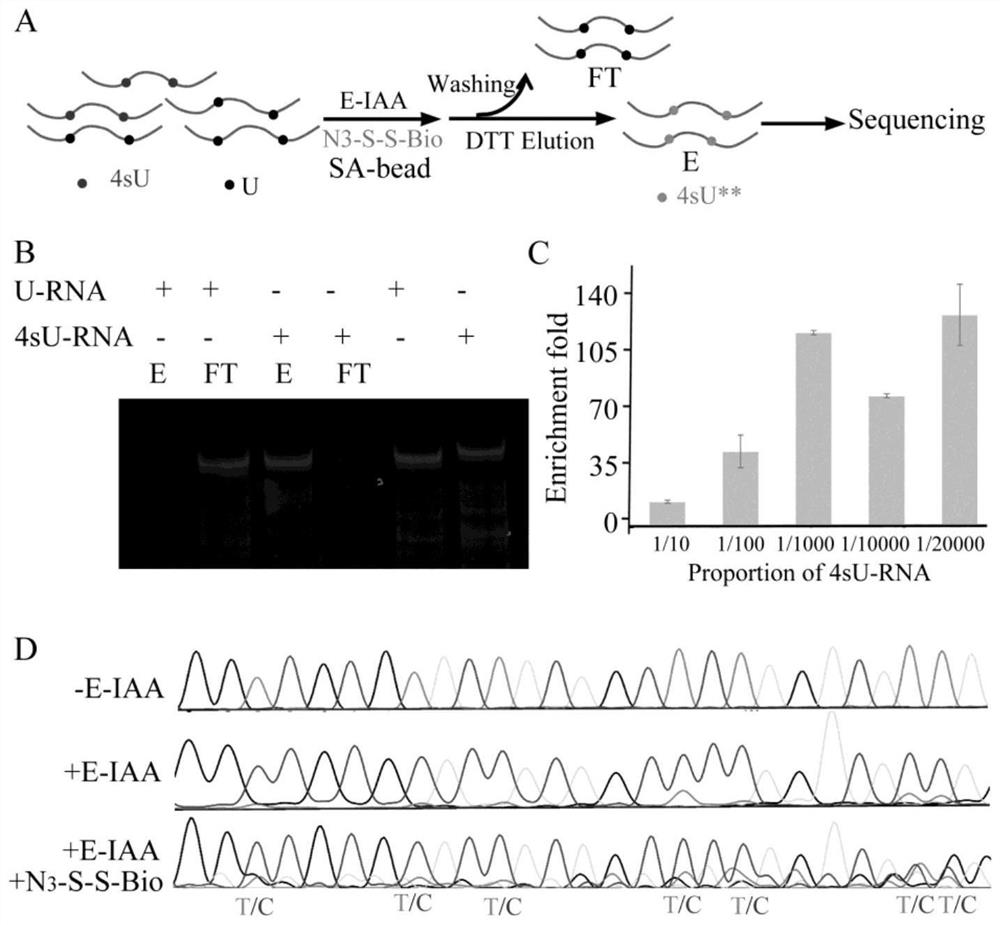
[0042] After enrichment of low-abundance 4sU-RNA by using the E-IAA described in the present invention to 4-mercaptouracil bifunctional chemical labeling, single-base resolution can still be performed.
[0043] RNAs containing 4-mercaptouracil and uracil were transcribed in vitro, and the sequences used to transcribe uracil RNA were q-pcr-U-cDNA mult-A and q-pcr-U-cDNAT7. The two RNAs were mixed at a ratio of 1 / 10, 1 / 100, 1 / 1000, 1 / 10000 and 1 / 20000, respectively, and the mixed samples were subjected to E-IAA reaction under standard conditions. Next, the purified RNA was subjected to click chemistry with an azide compound with disulfide bond biotin, so that 4sU-RNA was biotinylated. Streptavidin magnetic beads were used to enrich the RNA, and finally the DTT eluent was used to release the RNA from the magnetic beads. By calculating the ratio of the two RNAs after enrichment, the enrichment factor of 4sU-RNA by this method can be obtained. The sequences used for reverse trans...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com