PDA (at) CRISPR/Cas9/sgHMGA2 NPs complex, preparation method and application
Technology of a complex, sghmga2, applied in the biological field
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
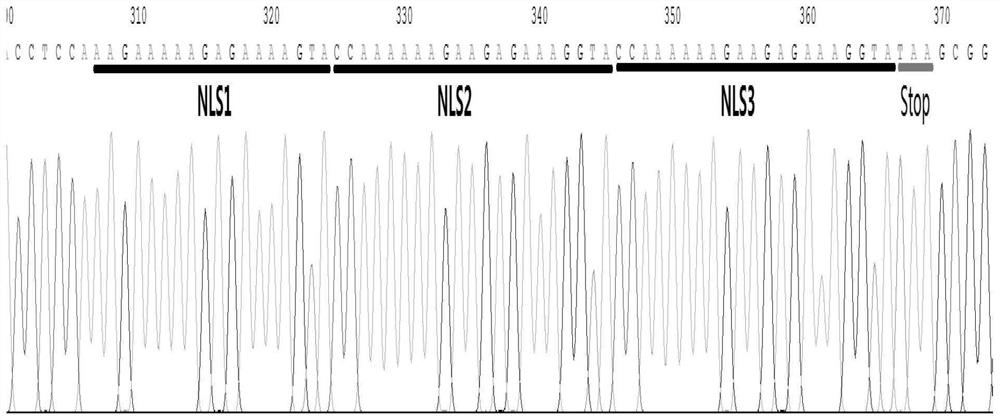
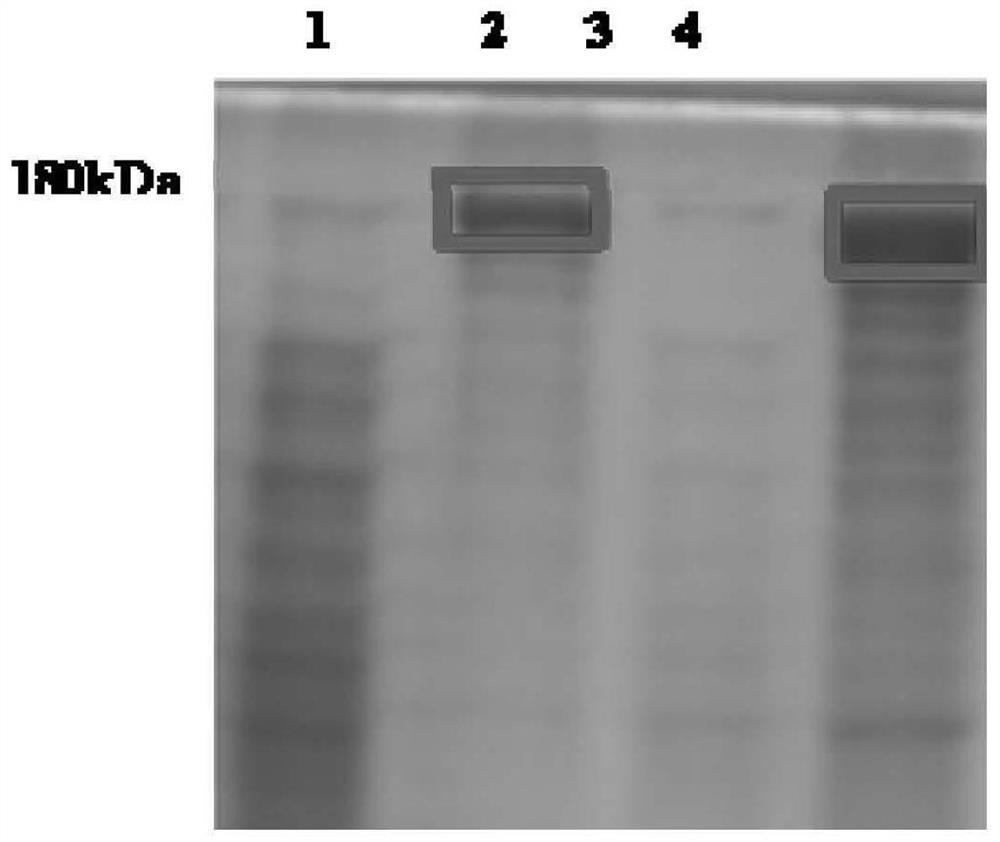
[0064] CRISPR / Cas9 -3NLS Preparation of endonuclease: In the Cas9 coding region of the backbone vector pET28a / Cas9-Cys (Addgene, Plasmid 53261) (before the nucleotide sequence encoding the SV40 nuclear localization signal sequence (NLS) (PKKKRKV)) additionally introduce two coding The nucleotide sequence of SV40 NLS, finally obtained the recombinant prokaryotic expression vector containing the nucleotide sequence of 3 SV40 NLS, named pET28a / Cas9-N3. The specific operation steps are as follows: according to the pET28a / Cas9-Cys vector sequence, Design specific primers N3-F and N3-R as shown in SEQ ID NO.:1, SEQ ID NO.:2: wherein the 5' end of N3-F contains a nucleotide sequence that can encode an NLS, N3-R The 5' end of contains nucleotide sequences encoding two NLSs. Use the above N3-F / -R primers and high-fidelity enzyme POD-plus-Neo (1U / ul) to perform PCR amplification on 100ng substrate pET28a / Cas9-Cys (conditions are as follows), purify and recover after agarose gel electro...
Embodiment 2
[0066]sgHMGA2, CRISPR sgRNA targeting HMGA2: CRISPR / Cas9 as shown in SEQ ID NO.:4, SEQ ID NO.:5 and SEQ ID NO.:6 was designed for the HMGA2 gene fragment shown in SEQ ID NO.:3 Target sequence, followed by synthesis of the linkers shown in SEQ ID NO.:7, SEQ ID NO.:8, SEQ ID NO.:9, SEQ ID NO.:10 and SEQ ID NO.:11, SEQ ID NO.:12 The target sequence and its complementary sequence were annealed to obtain three CRISPR sgRNA double-stranded DNA fragments as insert fragments, which were recombined with the DR274 vector treated with endonuclease (preferably BbsI endonuclease) to form three Recombinant prokaryotic expression plasmids targeting different sites of the HMGA2 gene: DR274-sgHMGA2-1, DR274-sgHMGA2-2 and DR274-sgHMGA2-3; the above three plasmids were digested with restriction endonuclease (DraI), The digestion product containing the T7 promoter and the CRISPR sgRNA targeting HMGA2 was recovered by electrophoresis and used as a template. After in vitro transcription and extract...
Embodiment 3
[0068] Preparation method of PDA@CRISPR / Cas9 / sgHMGA2 NPs complex:
[0069] S1, CRISPR / Cas9 -3NLS preparation of
[0070] S1.1. According to the pET28a / Cas9-Cys carrier sequence, design specific primers N3-F as shown in SEQ ID NO.:1, and design specific primers N3-R and N3-F as shown in SEQ ID NO.:2 The 5' end of N3-R contains a nucleotide sequence that can encode one NLS, and the 5' end of N3-R contains a nucleotide sequence that can encode two NLSs;
[0071] S1.2. Use N3-F and N3-R primers and high-fidelity enzyme POD-plus-Neo to perform PCR amplification on the substrate pET28a / Cas9-Cys;
[0072] S1.3. Purify and recover the target PCR product after agarose gel electrophoresis;
[0073] S1.4. After phosphorylation and self-ligation, the PCR product was subcloned into Escherichia coli DH5α, and then colony PCR and sequencing were performed to identify and screen the vector containing the nucleotide sequences encoding three NLSs;
[0074] S1.5. Using competent cell Rosetta...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com