A kind of nucleic acid nanostructure probe and its preparation method and application
A nucleic acid nano-probe technology, applied in biochemical equipment and methods, DNA preparation, chemical instruments and methods, etc., to achieve low detection limit, short synthesis time, and reduced loss efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0083] In Vitro Recognition of APE1 Enzyme Using Nucleic Acid Nanoprobes
[0084] (1) Synthetic probes
[0085] (1) Dilute the seven DNA single strands P1, P2, P3, P4, P5-CY5, P6-BHQ3, and P7-HE to 25 μM with enzyme-free water, and vortex for 10 seconds to fully mix the single strands.
[0086] (2) To establish a 50 μL system, add 31 μL enzyme-free water and 5 μL 10×TAE buffer-Mg 2+ , 2 μL P1, 2 μL P2, 2 μL P3, 2 μL P4, 2 μL P5-CY5, 2 μL P6-BHQ3, 2 μL P7-HE; add the sample on the side wall of the centrifuge tube, after adding the sample, centrifuge and mix.
[0087] (3) The PCR annealing procedure was used to combine the seven DNA single strands into a three-dimensional nucleic acid nanostructure. The PCR setting program is as follows: 80°C, 10min; 4°C, 30min. Store at 4°C until use.
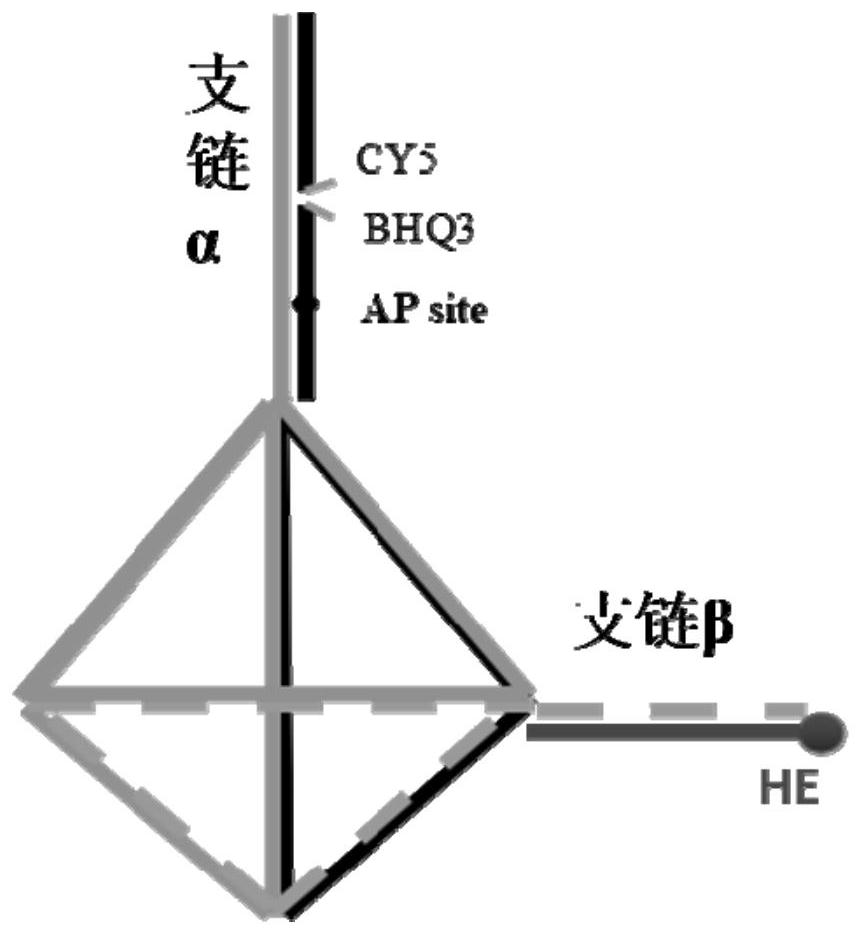
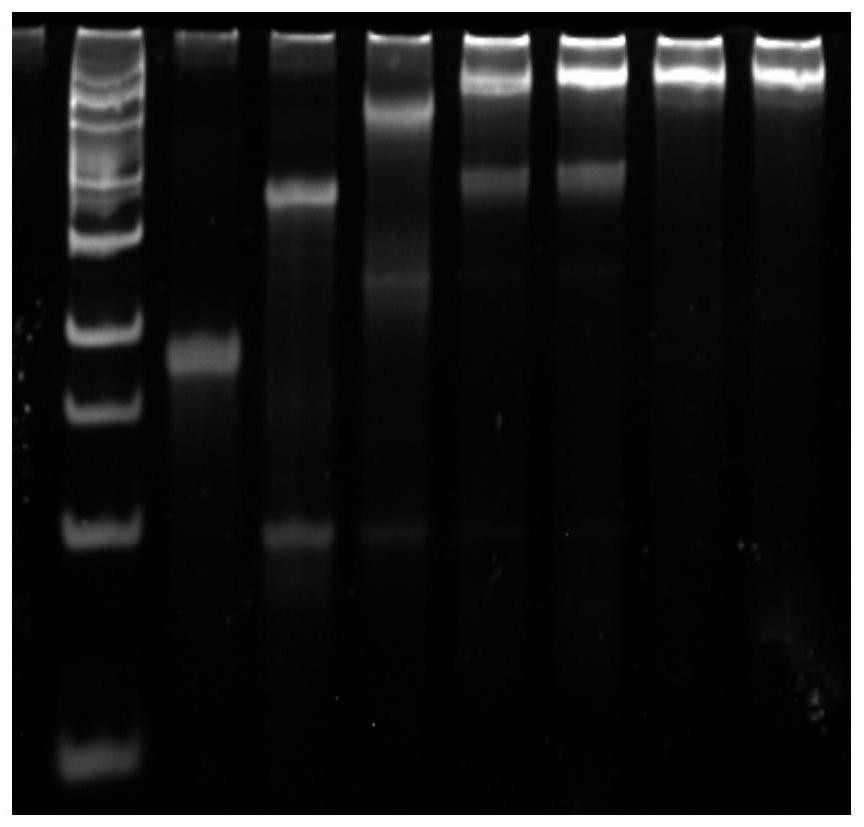
[0088] A schematic diagram of a nucleic acid nanostructure such as figure 1 Shown; Use polyacrylamide gel electrophoresis to the result that the probe synthesized by the embodiment of the p...
Embodiment 2
[0092] In vitro recognition of superoxide anion O using nucleic acid nanoprobes 2 · -
[0093] (1) Synthetic probes
[0094] Method is with embodiment 1 (one)
[0095] (2) In vitro identification and results
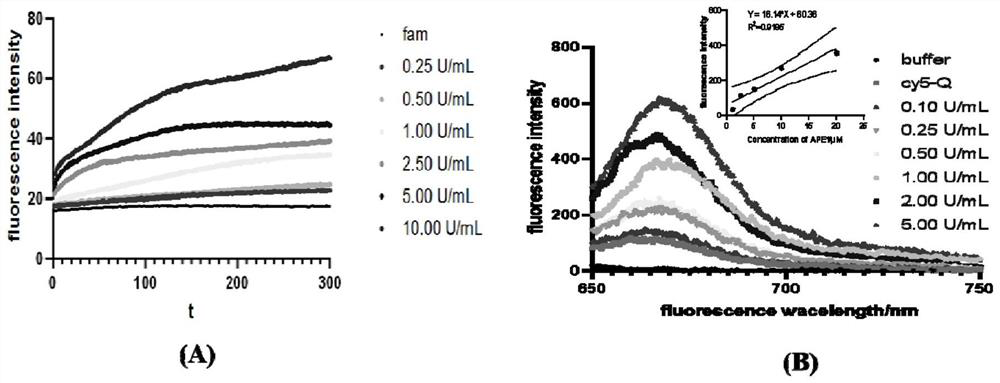
[0096] The synthesized probe was reacted with different concentrations of xanthine / xanthine oxidase in PBS buffer solution, and the fluorescence response intensity was between Figure 4 shown in . The experiment was performed in a fluorimeter. 1mol of xanthine and 1U of xanthine oxidase can generate 0.33mol of superoxide anion. The initial velocity of the fluorescence-concentration curve showed a good linear relationship between the probe and the concentration of superoxide anion in the range of 0.33-133.33mol / L. The detection limit was determined to be 0.33mol / L.
Embodiment 3
[0098] Application of nucleic acid nanoprobes to study APE1 expression and redox levels in cells exposed to PMA
[0099] PMA (phorbol ester) is a stimulator of reactive oxygen species. Cells exposed to PMA will lead to an increase in the content of reactive oxygen species and an imbalance in redox levels.
[0100] (1) Synthetic probes
[0101] Method is with embodiment 1 (one)
[0102] (2) Cell fluorescence experiment
[0103] (1) Cell receiving dish: Take out the tested cells from the constant temperature incubator, wash the cells with 1×PBS, draw 1ml of fresh complete medium with a pipette gun and repeatedly suck and blow the bottom of the culture flask, and mix the cells evenly. Pipette 1ml of the cell mixture into a 1.5ml EP tube and centrifuge at 1000rmp for 3min. Discard the supernatant after centrifugation, blow and mix the cells with fresh complete medium, pipette 200 μL of the uniform mixture into the groove of the confocal culture dish, dilute the cell suspension ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com