Cyclic RNA (Ribonucleic Acid) biomarker for breast cancer and application of circular RNA biomarker
A breast cancer, annular technology, applied in the field of breast cancer diagnosis, can solve problems such as lack of detection of breast cancer, and achieve the effects of improving the utilization efficiency of medical resources, improving the efficiency of diagnosis, and stabilizing the structure.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
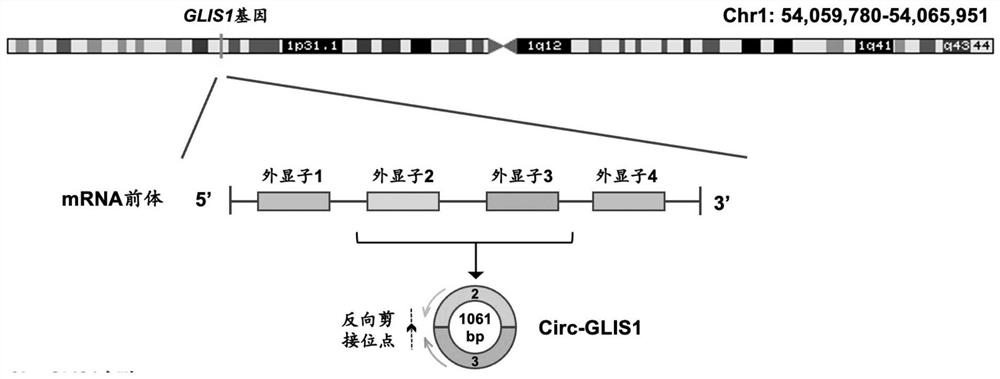
[0035] Example 1 Preparation method and structure verification of circular RNA circ-GLIS1
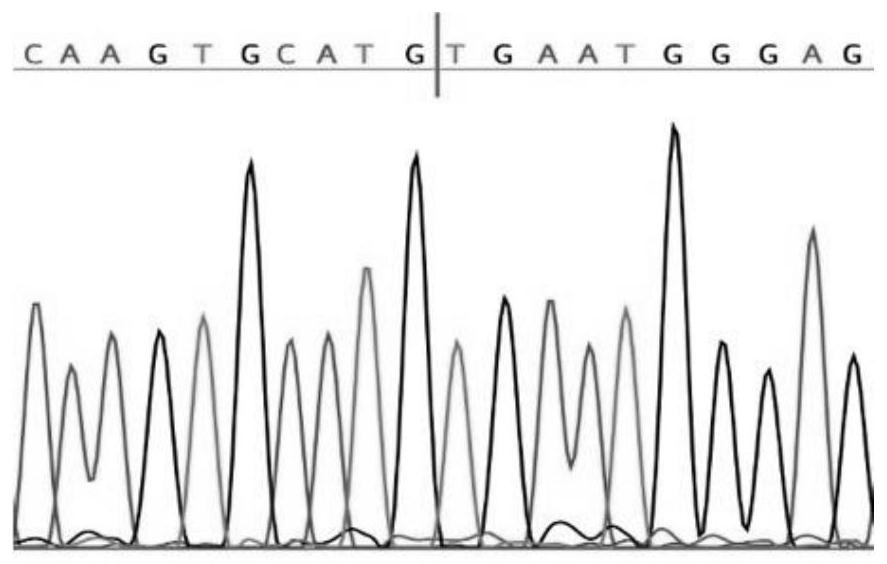
[0036] Circ-GLIS1 (hsa_circ_0002079, chr1:54059780-54065951), formed by reverse splicing of the second and third exons of the gene GLIS1 (1p32.3), has a length of 1061 bases. The structural characteristics and sequence information of Circ-GLIS1 are as follows: figure 1 and shown in SEQ No.1.
[0037] Quantitative detection method of circular RNA circ-GLIS1:
[0038] 1. RNA extraction: The total RNA of breast tumor cell MCF-7 was extracted with a total RNA extraction kit, and the purity and concentration of total RNA were detected with a NanoDrop3000 micro-nucleic acid analyzer.
[0039] 2. Reverse transcription: perform reverse transcription to obtain a cDNA template. The reverse transcription reaction steps are: template RNA 500ng, random hexamer primer 1μL, RNase-free water to a total volume of 12μL, mix and store at 70°C Incubate for 5 minutes; then add 4 μL of 5× reaction buffer,...
Embodiment 2
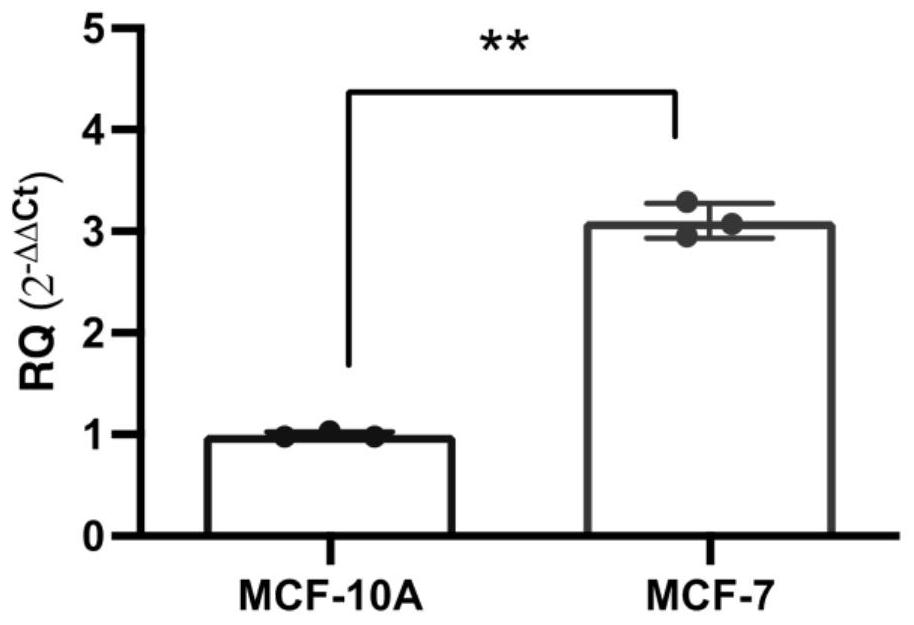
[0043] Example 2 Detection of circular RNA Circ-GLIS1 expression in breast tumor cells and normal breast cells
[0044] Fluorescent quantitative PCR (qPCR) was used to detect the expression of circ-GLIS1 in breast tumor cell MCF-7 and breast normal cell MCF-10A (purchased from the Cell Bank of the Chinese Academy of Sciences, Shanghai), and found that circ-GLIS1 was expressed in breast tumor cells. The expression was significantly higher than that in normal cells, indicating that circ-GLIS1 can act as a tumor-promoting factor and a potential therapeutic target for breast cancer.
[0045] Circular RNA circ-GLIS1 detection method: Total RNA of breast tumor cell MCF-7 and normal breast cell MCF-10A was extracted using a total RNA extraction kit, and the purity and concentration of total RNA were detected with a NanoDrop3000 micro-nucleic acid analyzer. Then, use the aforementioned method to perform reverse transcription to obtain cDNA templates for qPCR detection. The experimenta...
Embodiment 3
[0046] Example 3 Construction of Circ-GLIS1 overexpression vector and its effect on promoting the proliferation and invasion of breast cancer cells
[0047] The pCD5-ciR vector overexpressing circ-GLIS1 was constructed (Guangzhou Jisai Biotechnology Co., Ltd.), the full-length 1061bp sequence of hsa_circ_0002079 was amplified by PCR, and EcoRI and BamHI double enzyme digestion ligated it into pCD5-ciR. The pCD5-ciR vector structure is as follows Figure 4 shown.
[0048] Breast tumor cells were transfected with pCD5-ciR vector overexpressing circ-GLIS1, and the effect of overexpressing circ-GLIS1 on the proliferation and invasion ability of breast tumor cells was observed. Test steps: ① Inoculate MCF-7 cells in a 12-well plate, and transfect when they grow to 70% confluence; ② Add 1 μg circ-GLIS1 pCD5-ciR vector and 2 μL P3000 reagent mixture to 50 μL serum-free medium ③Add 2 μL Lipofectamine 3000 to 50 μL serum-free medium; ④Mix the diluted overexpression vector and Lipofec...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com