Early detection of liver cancer based on F12 gene methylation
A methylation and gene technology, applied in the determination/inspection of microorganisms, biochemical equipment and methods, DNA/RNA fragments, etc., can solve the problems of unfavorable technical transformation, cumbersome steps, high cost, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
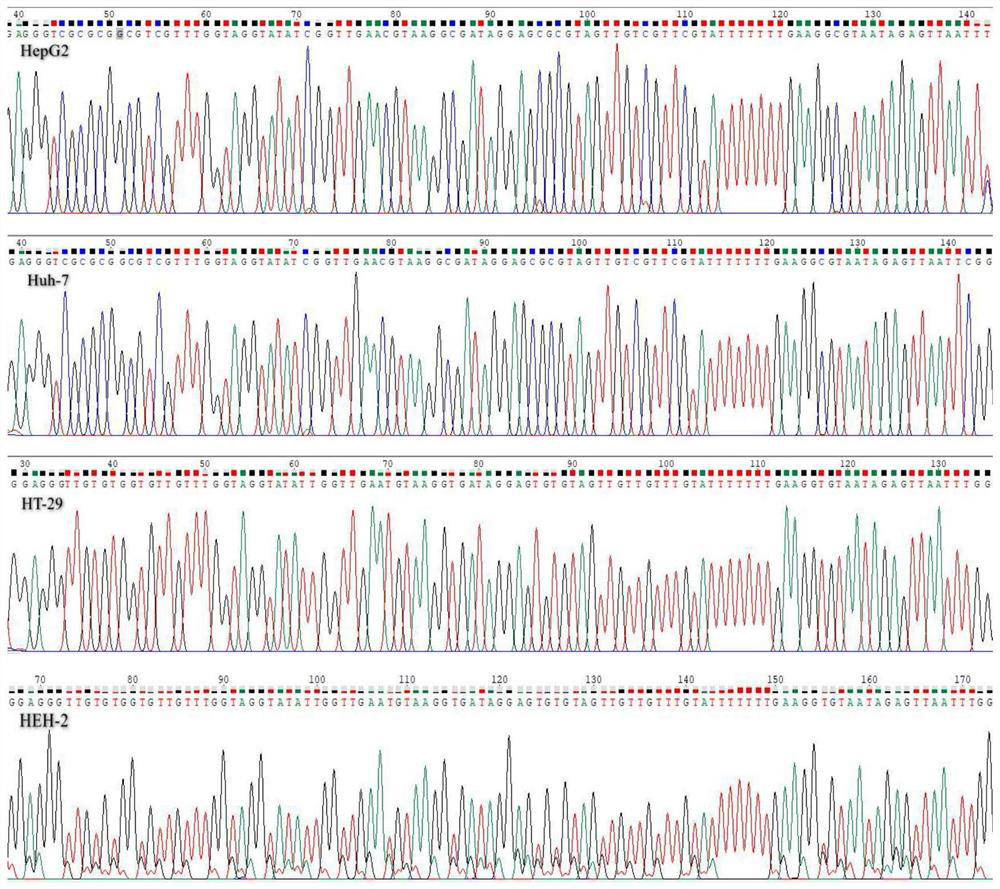
[0101] Example 1: Comparison of methylation levels of F12 in DNA of different cell lines
[0102] The inventors of the present invention based on the data analysis results of the whole blood methylation chips of paracancerous tissues of different cancer types and healthy people in the TCGA and GEO databases, and screened out liver (paracancerous) tissue-specific hypermethylated F12 as liver tissue-specific Methylation candidate markers. Since the methylation chip mainly detects the average methylation level of a sequence containing several CpG islands in the sample, in order to determine the methylation sites suitable for designing detection primer probes, the inventors used the methylation generation sequencing method to The target CpG sites of the above target genes were verified for methylation level one by one.
[0103] According to the analysis results of the methylation chip, the amplification and sequencing primers of the F12 candidate region were designed. The specifi...
Embodiment 2
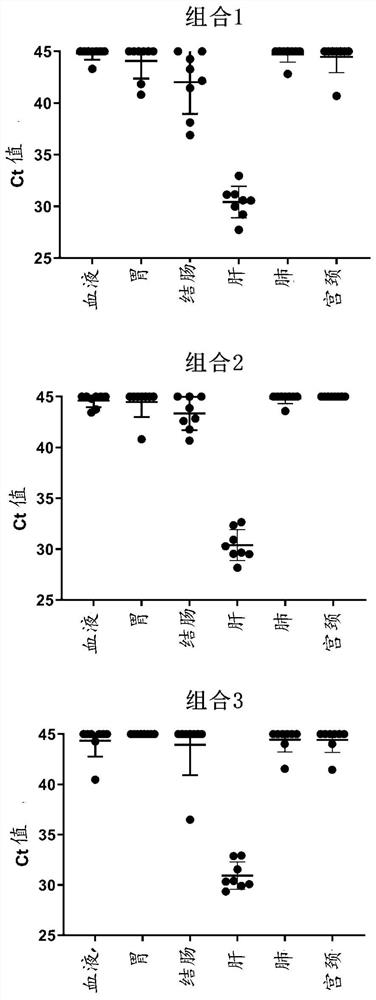
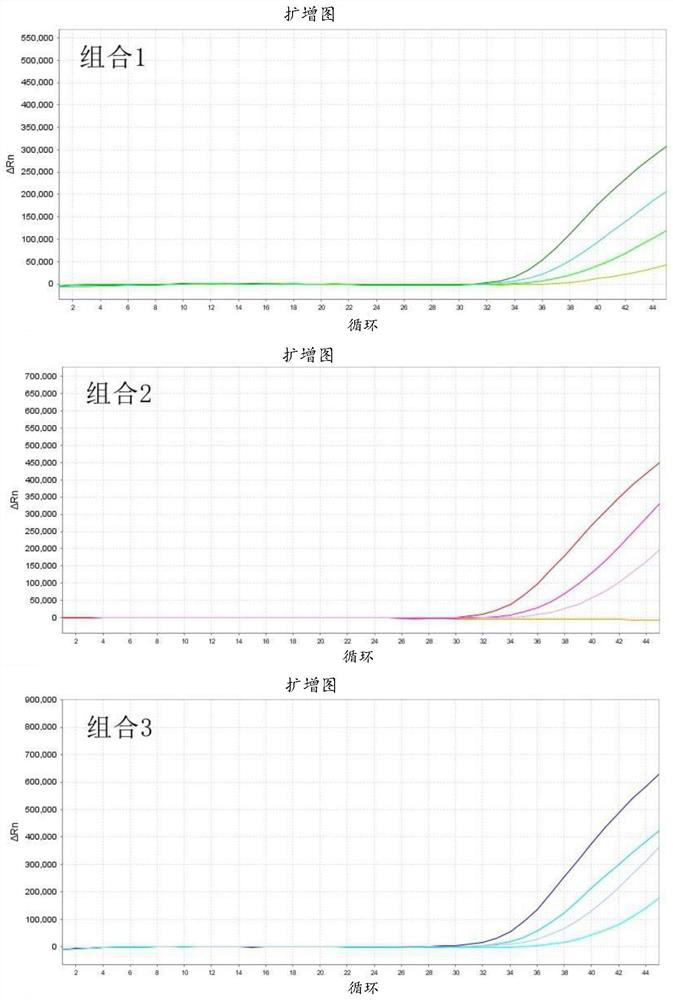
[0127] Example 2: Tissue-specific study of F12 methylation
[0128] Since most of the cell lines used in Example 1 are cancer cells, compared with non-cancerous cells, the cancer cells themselves may have undergone abnormal methylation in some regions of the genome. Therefore, in the experimental process of the present invention, 3 pairs of primer-probe combinations were designed for several CpG sites found in the sequence of the F12 gene promoter region for subsequent fluorescent quantitative PCR verification. The objects of verification are tissue DNA samples from different organ sources. Table 4 lists the target regions of the F12 gene selected in the experimental process of the present invention.
[0129] Table 4 The sequence of the target region of F12 gene
[0130]
[0131] The F12 forward primers include:
[0132] SEQ ID NO. 7F12-F1: TTGGTAGAGCGTGGTTTCGG
[0133] SEQ ID NO. 8F12-F2: CGTTTGGTAGGTATATCGGTTG
[0134] SEQ ID NO. 9F12-F3: GGACGTCGGGGTTTTAAGTT
[013...
Embodiment 3F12
[0167] Example 3 Analysis Sensitivity Verification of F12 Methylated Targets
[0168] Plasma contains a small amount of free DNA (cfDNA), which originates from different tissues in the body and is released into the blood through apoptosis, necrosis or extracellular secretion. The concentration of cfDNA in healthy people is about 10ng / ml in the human body, and the cfDNA derived from liver tissue accounts for about 1-2% of the total cfDNA. Since cfDNA has a short half-life (0.5-2h) and is prone to degradation, it is necessary to ensure that the detection limit of F12 methylated targets is less than 1%. The inventors used the following experiments to evaluate the minimum detection limit of the three F12 primer-probe combinations on the F12 positive target in the background of 10 ng of negative template DNA.
[0169] The detection primers and probes for each gene are as follows:
[0170] F12 forward primers include:
[0171] SEQ ID NO. 7F12-F1: TTGGTAGAGCGTGGTTTCGG
[0172] SE...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com