MNP core primer combination for molecular identification of kiwi fruit varieties and application of MNP core primer combination
A technology of molecular identification and primer combination of varieties, applied in the field of molecular biology, to ensure production safety and speed up the breeding cycle
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] Screening and Validation of MNP Core Marker Primer Combinations for Molecular Identification of Kiwifruit DNA Varieties
[0024] (1) Screening of kiwifruit MNP primer marker combinations
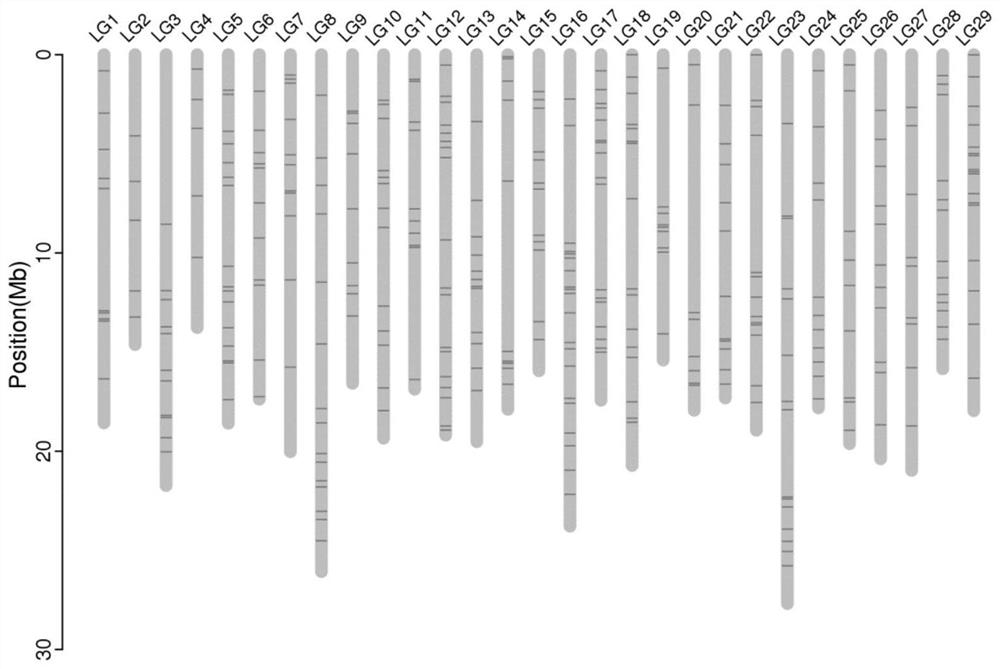
[0025] Taking the publicly released kiwifruit genome as a reference (http: / / eggplant.kazusa.or.jp / ), using the resequencing data of 76 kiwifruit genomes carried out by our team for comparative analysis, 347 MNP marker sites were screened ( figure 1 ), design primers (see Table 1 for primer details).
[0026] (2) Correctness analysis of kiwifruit MNP primer marker combination
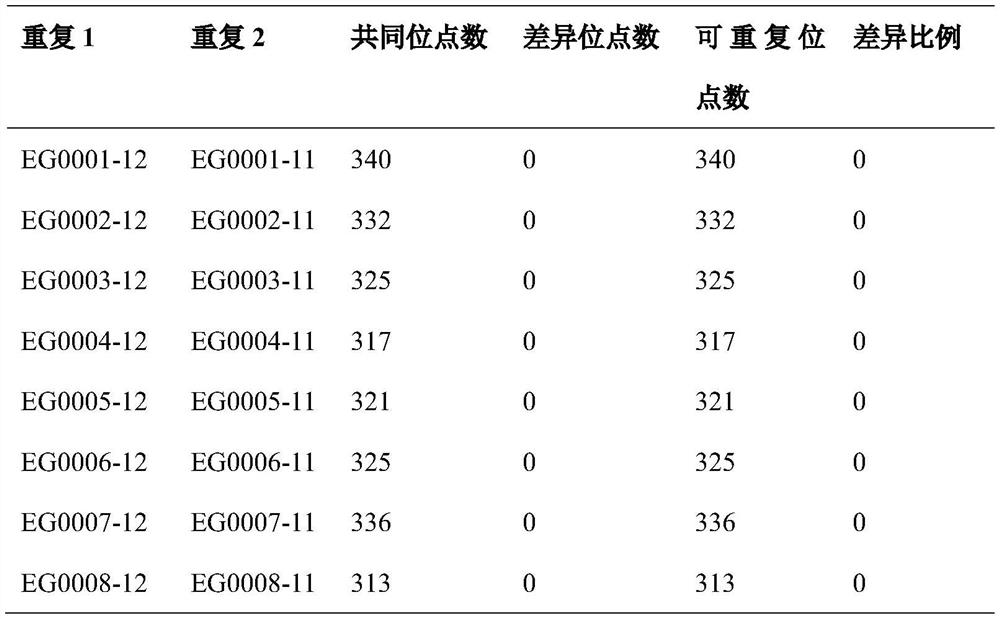
[0027] In order to test the accuracy of the kiwifruit MNP marker method, 8 samples were collected from the national kiwifruit resource garden, and a reproducibility experiment was carried out. The same DNA of each variety was constructed twice, and the two libraries were sequenced twice at different times. When comparing the first and second sequencing of different batches of libraries, the number of differenc...
Embodiment 2
[0033] Construction of standard DNA fingerprint of kiwifruit with MNP core marker primer combination for molecular identification of kiwifruit DNA varieties
[0034]1. DNA extraction and quality assessment
[0035] Genomic DNA of all kiwifruit varieties used to construct the kiwifruit fingerprint database was extracted by conventional CTAB method, and the quality of DNA was detected by agarose gel and ultraviolet spectrophotometer after RNase treatment. If the DNA band is single, there is no obvious degradation, the UV spectrophotometer detects that the A260 / 280 is between 1.8-2.0, and the A260 / 230 is between 1.8-2.0, there is no obvious light absorption at 270nm, and the DNA concentration is greater than 50ng / ul, it means Genomic DNA meets the relevant quality requirements and can be used for subsequent experiments.
[0036] 2. MNP marker detection
[0037] Using 347 synthetic kiwifruit MNP-labeled primers, multiplex amplification, next-generation high-throughput sequencing...
Embodiment 3
[0041] Authenticity of kiwifruit varieties detected by MNP-labeled primer combination for molecular identification of kiwifruit DNA varieties
[0042] 1. Determination of the discrimination degree of kiwifruit MNP primer marker varieties and the determination of the authenticity test results of kiwifruit varieties
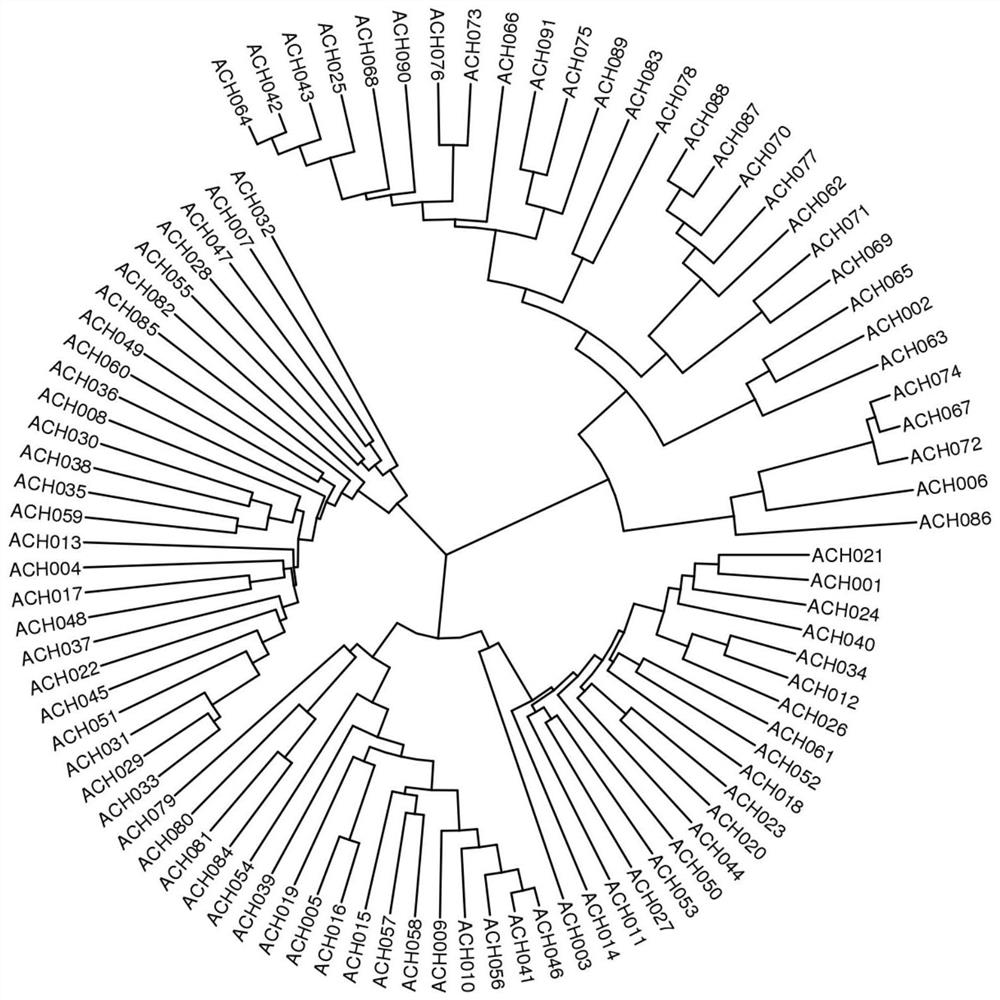
[0043] The 91 kiwifruit samples in Example 2 were pairwise compared to analyze the differences of their MNP marker sites, and a total of 4095 pairs of comparison results were obtained, the average number of MNP marker differences between any varieties was 89.22, and the genetic distance difference of 4095 pairs of samples Between 0.30% and 82.26%, the average difference is 47.85%, and no difference is 0 in pairwise comparison. Using the developed marker detection method, 100% of kiwifruit varieties could be distinguished ( figure 2 ), indicating that the developed kiwifruit MNP markers have high polymorphism and strong variety discrimination. Therefore, the 347 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com