Primer and kit for detecting CGG repetition number of FMR1 gene
A kit and repeat number technology, applied in the field of biological diagnosis, can solve the problems of high experimental requirements, complex system and high cost, and achieve the effects of improving detection efficiency and quick and easy method.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
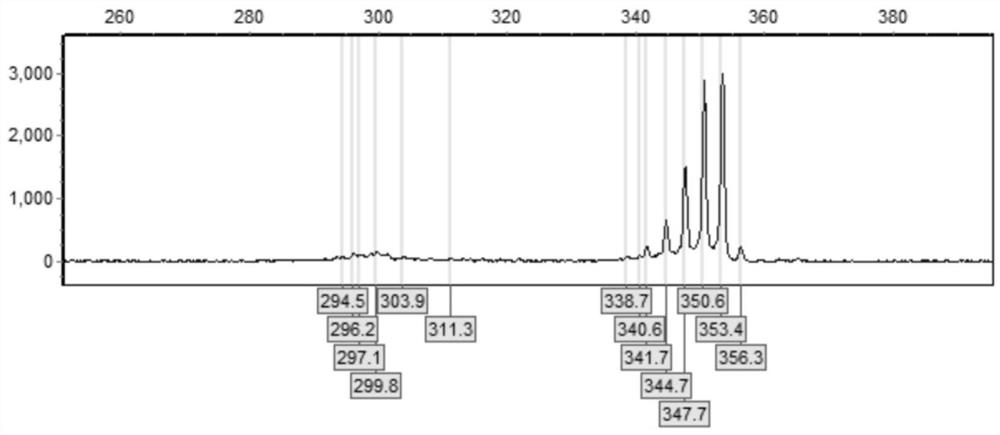
[0034] Example 1 Design and detection of primers for detection of CGG repeat number of FMR1 gene
[0035] (1) Design primers F and R for amplifying and detecting target fragments at the two ends of the FMR1 gene CGG repeat sequence; the sequence of the upstream primer F for detecting the CGG repeat number is as shown in SEQ ID NO: 1, and its 5' FAM fluorescence at the end; the sequence of the downstream primer R is shown in SEQ ID NO: 2; the specific table below:
[0036] serial number Primer (5'-3') SEQ ID NO:1 FAM-AGCCCCGCACTTCCCACCACCAGCTCCTCCA SEQ ID NO:2 GCTCAGCTCCGTTTCGGTTTCACTTCCGGT
[0037] (2) Reaction system:
[0038] Mix1:
[0039] Reagent System (μL) Water 1.8 1mM dATP 0.75 1mM dCTP 0.75 1mM dTTP 0.75 5mM 7-deaza-ATP 0.05 10uM SEQ1(FMR1-F) 0.2 10uM SEQ2(FMR1-R) 0.2 GC-Rich resolution solution 3.5 (individually added)
[0040] Mix2:
Embodiment 2
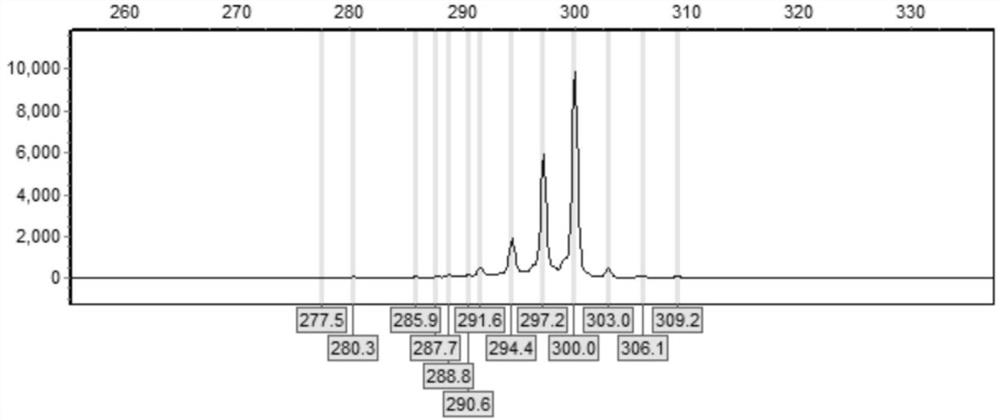
[0050]Example 2 Detection of CGG Repeat Numbers in Normal Male Samples
[0051] (1) DNA extraction: Peripheral blood was collected with the consent of the subject or the knowledge of his guardian. A commercially available genomic DNA extraction kit was used to extract genomic DNA from peripheral blood samples.
[0052] (2) Use the primers of Example 1 and kits thereof to prepare a PCR reaction system, carry out PCR amplification to the sample FMR1 gene CGG repeat sequence, and the PCR reaction system includes the first PCR reaction solution (Mix1) and the second PCR reaction solution ( Mix2), the components of the first PCR reaction solution (Mix1) are: water: 1.8 μL; 1mM dATP: 0.75 μL; 1mM dTTP: 0.75 μL; 1mMdCTP: 0.75 μL; 5mM 7-deaza-ATP: 0.05 μL; 10μM F: 0.2μL; 10μM R: 0.2μL; GC-Richresolution solution: 3.5μL;
[0053] The components of the second PCR reaction solution (Mix2) are: water: 0.8 μL; 5×GC-Rich Reaction Buffer: 2 μL; GC-Rich Enzyme Mix: 2 μL;
[0054] After the...
Embodiment 3
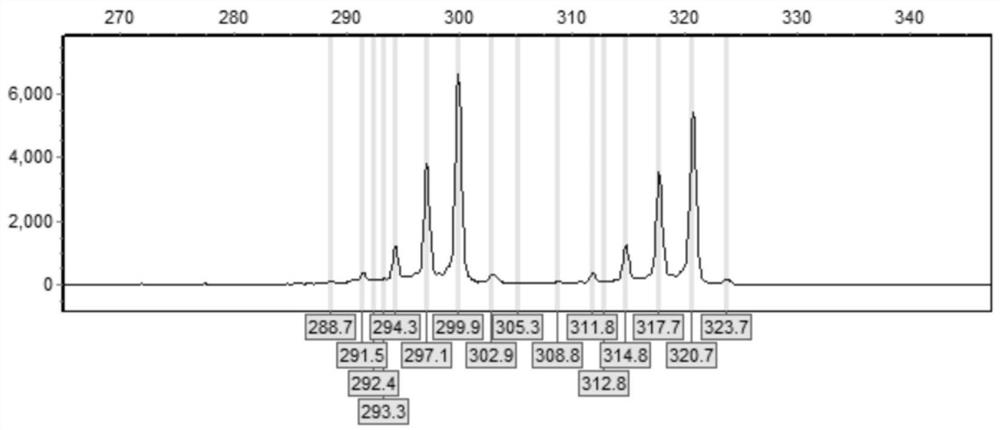
[0058] Example 3 Detection of CGG Repeat Numbers in Normal Female Samples
[0059] (1) DNA extraction: Peripheral blood was collected with the consent of the subject or the knowledge of his guardian. A commercially available genomic DNA extraction kit was used to extract genomic DNA from peripheral blood samples.
[0060] (2) Use the primers of Example 1 and kits thereof to prepare a PCR reaction system, carry out PCR amplification to the sample FMR1 gene CGG repeat sequence, and the PCR reaction system includes the first PCR reaction solution (Mix1) and the second PCR reaction solution ( Mix2), the components of the first PCR reaction solution (Mix1) are: water: 1.8 μL; 1mM dATP: 0.75 μL; 1mM dTTP: 0.75 μL; 1mMdCTP: 0.75 μL; 5mM 7-deaza-ATP: 0.05 μL; 10μM F: 0.2μL; 10μM R: 0.2μL; GC-Richresolution solution: 3.5μL;
[0061] The components of the second PCR reaction solution (Mix2) are: water: 0.8 μL; 5×GC-Rich Reaction Buffer: 2 μL; GC-Rich Enzyme Mix: 2 μL;
[0062] After ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com