Molecular marker of F3 'H homologous gene and application thereof
A genome and primer combination technology, applied in the field of biological breeding, can solve problems such as the unfavorable commercial value of broccoli curds, and achieve the effects of reducing the scale of planting and identification, high throughput, and accurate data collection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
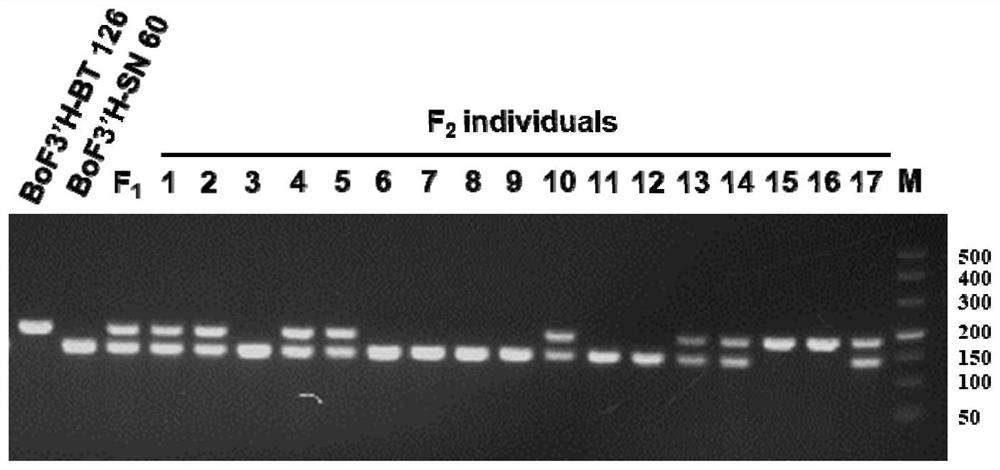
[0032] The purple curd broccoli material obtained in the field is the inbred line BT 126 as the female parent, the green curd broccoli material inbred line SN60 is the male parent (both BT126 and SN60 are from Shanghai Fengxian Zhuanghang Farm), and the female parent BT 126 is High anthocyanin content, the color of the curd is purple, and the male parent SN 60 has low anthocyanin content, and the color of the curd is green.
[0033] Through hybridization and selfing, 302 individual plants of the F2 isolated population were obtained, and the phenotype verification of the F2 population was carried out. The specific steps are as follows:
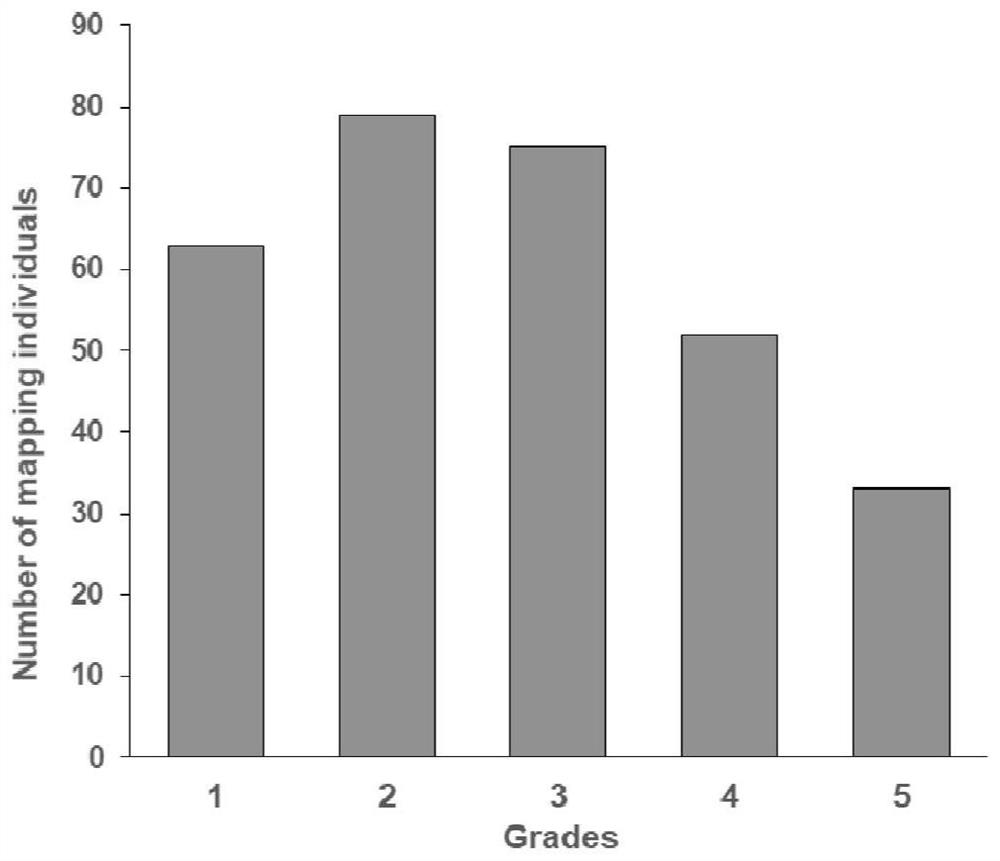
[0034] The F2 population plants were planted in a greenhouse, and the phenotypes of each individual plant were identified at least three times when the curds were mature. The curd color shows the color distribution from green to purple, divided into 5 grades, from 1 to 5: 1-green, 2-purple, 3-light purple, 4-slightly darker purple, and 5-purple...
Embodiment 2
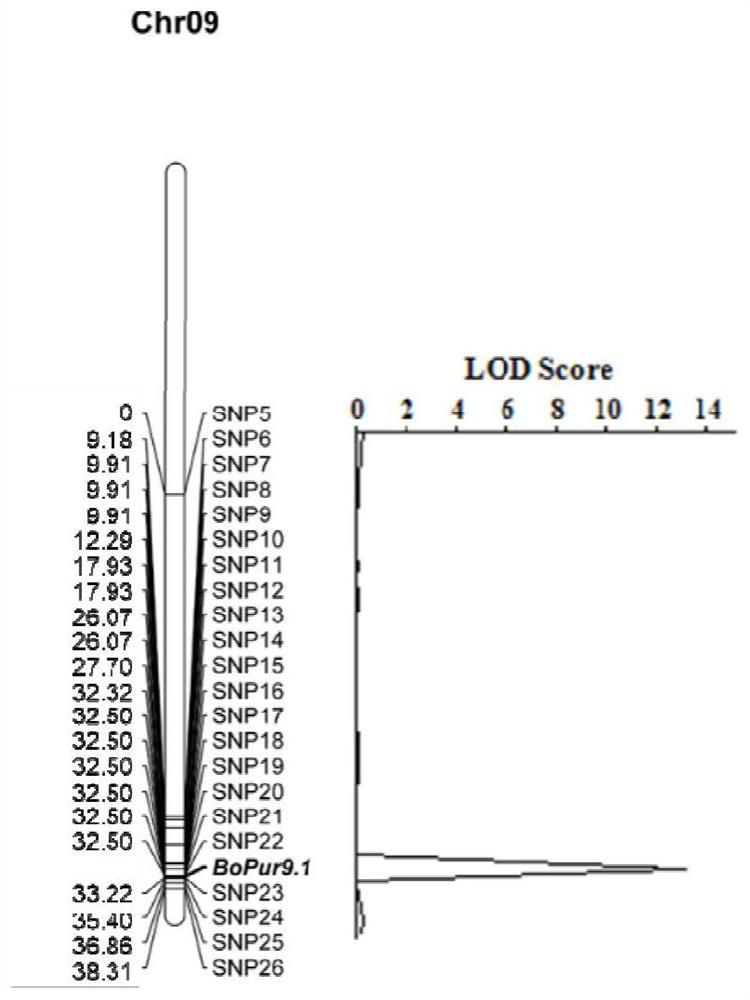
[0040] A pair of primers, IndBoF3'H-F and IndBoF3'H-R, were designed for the deletion of F3'H-SN 60 and the structural features of F3'H-BT 126 and F3'H-SN 60. 126 was amplified to yield a 187 bp fragment, and SN 60 was amplified to yield a 144 bp fragment.
[0041] SEQ NO.1: (IndBoF3'H-F) CACGGACATGCTTAGCACTTTAAT
[0042] SEQ NO.2: (IndBoF3'H-R) GCCGTAAACATGTTCTGTAATGGA
[0043] The primers were synthesized by Sangon Bioengineering (Shanghai) Co., Ltd.
Embodiment 3
[0045] 1. Using CTAB method, genomic DNA was extracted from broccoli leaves.
[0046] (1) Take an appropriate amount of fresh or frozen leaves of broccoli seedlings, put them into a 2ml centrifuge tube, add two steel balls, and grind them on a tissue grinder;
[0047] (2) Add 750 μL CTAB solution, shake to homogenize, and shake at 65°C for 0.5-1 h;
[0048] (3) Cool to room temperature, add 700 μL of chloroform:isoamyl alcohol (24:1, volume ratio) in a fume hood, and mix by hand and gently upside down for 15~20min;
[0049] (4) Centrifuge at 12000rpm for 12min, and take 400μL of supernatant into a new 1.5mL centrifuge tube;
[0050] (5) Add 2 volumes (800 μL) of ice ethanol to the supernatant, cover the centrifuge tube, shake it a few times, mix the ice ethanol and the supernatant well, and put it in a -20°C refrigerator Let stand for 20~30min;
[0051] (6) Centrifuge the standing centrifuge tube at 12,000 rpm for 5 minutes, then pour off the supernatant, add 500 μL of 75% ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com