Method for visually detecting Hangzhou white chrysanthemum leaf blight chaetomium globosum by using CRISPR/Cas12a system
A technology for Chaetomium globulus and leaf blight, which is applied in the fields of biology and chemistry, can solve the problems of expensive experimental equipment, low sensitivity, long time consumption, etc., and achieves the effects of user-friendliness, low price and high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0027] The technical solutions of the present invention will be further illustrated and described below through specific embodiments in conjunction with the accompanying drawings:
[0028] 1. Visual detection of C. globosum by CRISPR / Cas12a system
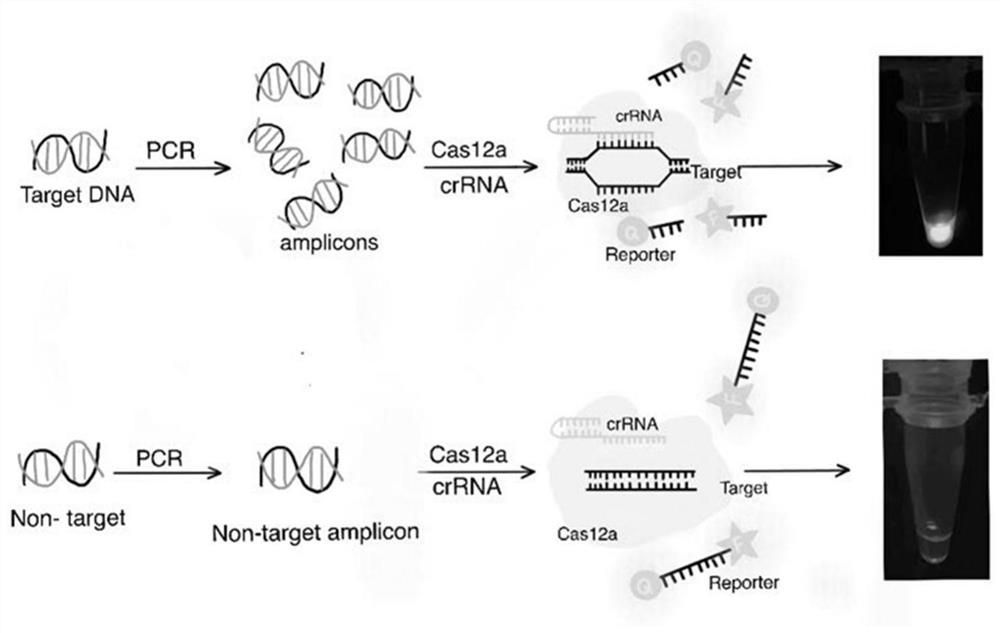
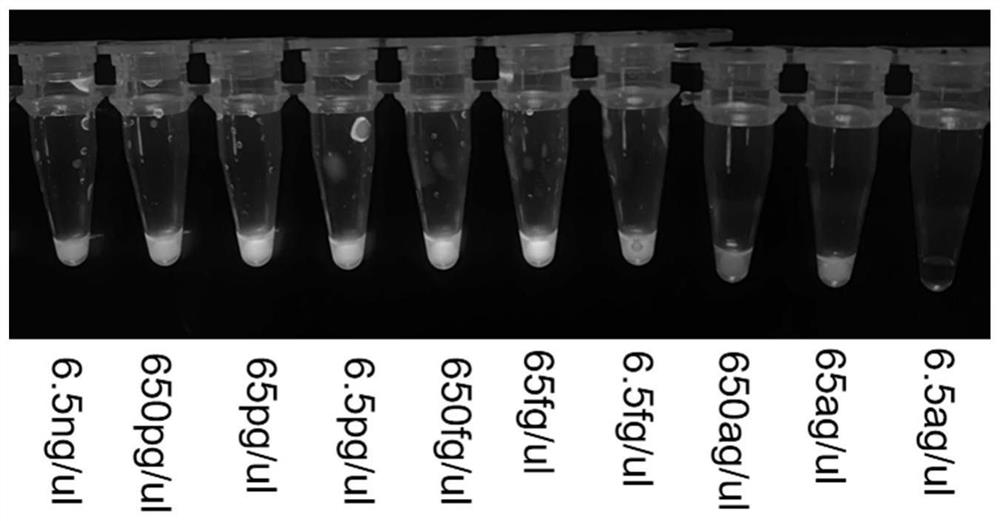
[0029] The actual samples were pretreated by thermal lysis method. Under sterile conditions, 1 mL of the sample homogenate was taken into a sterile centrifuge tube, centrifuged at 8000 × g for 2 min, and the supernatant was discarded; Centrifuge at ×g for 2 min, discard the supernatant; add 100 μL of sterile deionized water, boil at 100°C for 10 min; centrifuge the thermal lysis solution at 8000 × g for 2 min, and take the supernatant as the substrate for the subsequent amplification reaction (i.e. Target DNA ),like figure 1 for the specific operation steps. The PCR amplification technology is used to amplify the target fragment, and the primers used in the specific gene fragment system are SEQ ID No.1 and SEQ ID No.2.
[0030] ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com