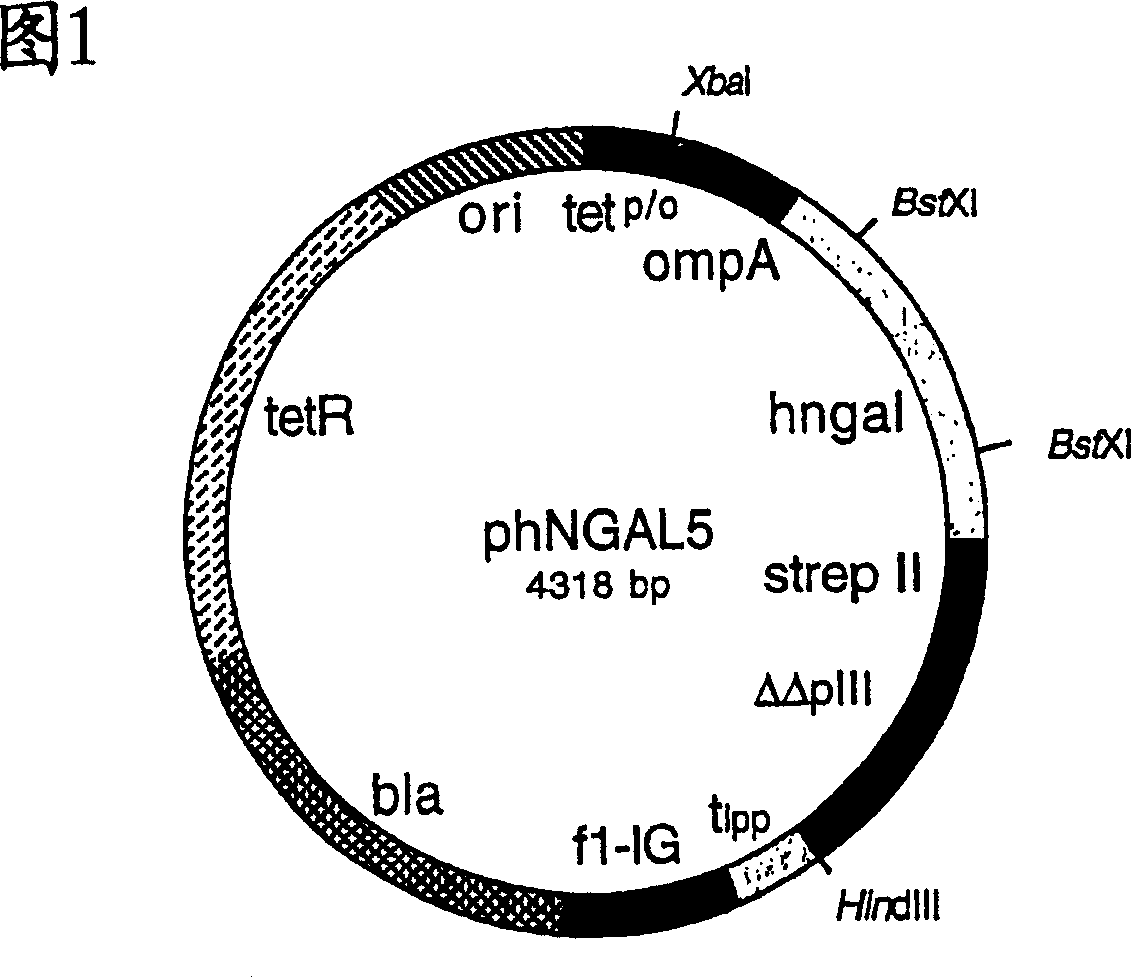
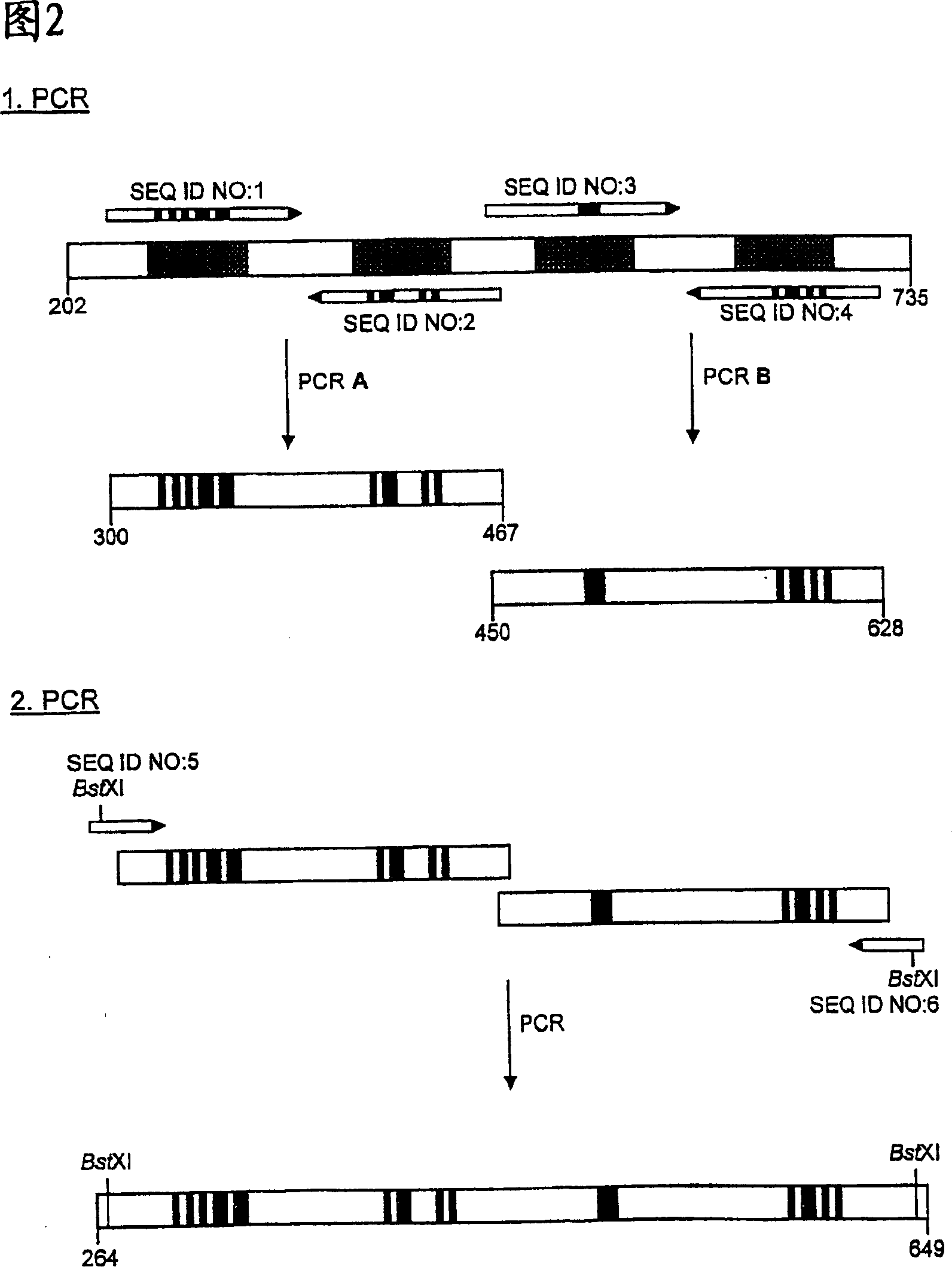
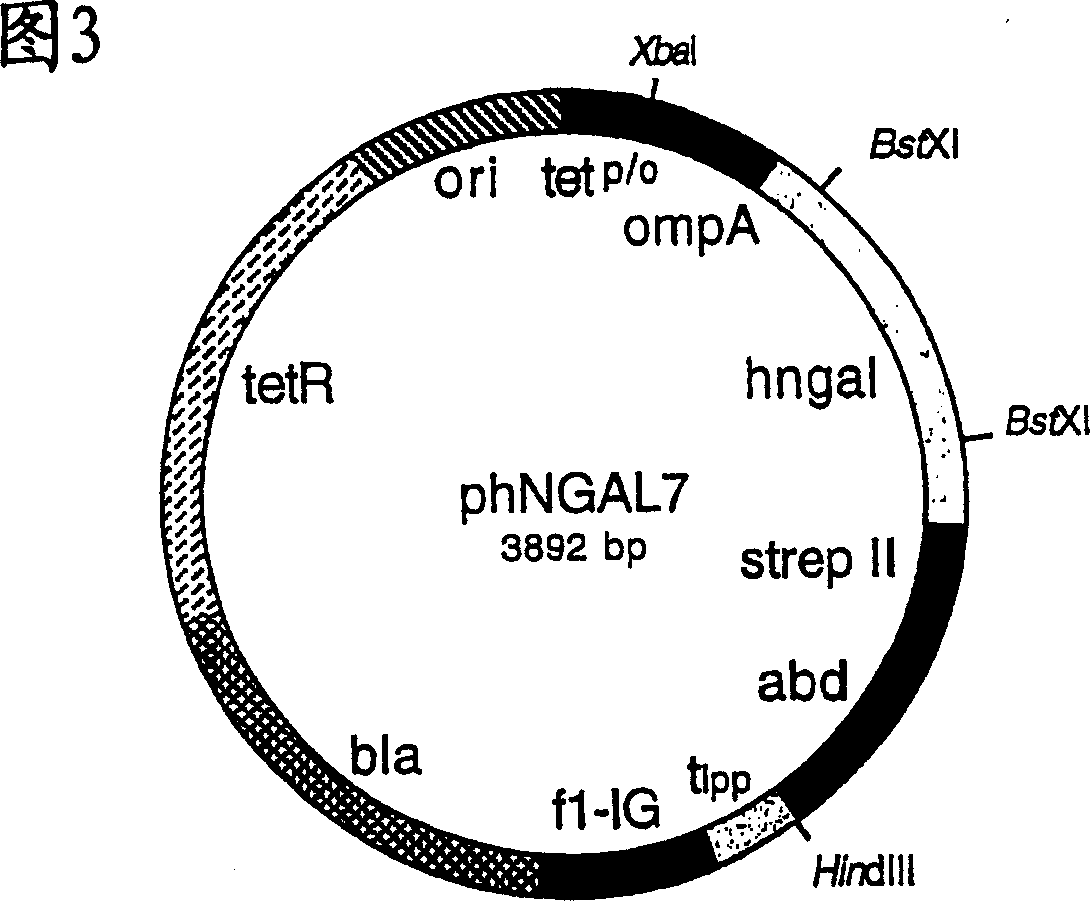
Muteins of human neutrophil gelatinase-associated lipocalin and related proteins
A neutrophil and mutant protein technology, applied in the field of human neutrophil gelatinase-related lipocalin and mutant proteins of related proteins
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0109] Example 1: Preparation of hNGAL mutant protein library
[0110] Unless otherwise indicated, genetic engineering methods known to those skilled in the art, such as those described by Sambrook et al. (supra) were used.
[0111] PCR was used in multiple steps according to Figure 2 to allow for concerted mutagenesis at all 20 selected amino acid positions in the four peptide loops of hNGAL. A PCR reaction of 100 μl volume was carried out in both first amplification steps, wherein 20 ng of phNGAL3 plasmid DNA was used as a template, and 50 pmol each of the corresponding primers (respectively SEQ ID NO: 1 and SEQ ID NO: 2 or SEQ ID NO: 1 and SEQ ID NO: 2 or SEQ ID NO: NO: 3 and SEQ ID NO: 4), these primers were synthesized by the traditional phosphoramidite method. Additionally, the reaction mixture contained 10 μl of 10xFaq buffer (100 mM Tris / HCl pH 9.0, 500 mM KCl, 15 mM MgCl 2 , 1% (v / v) Triton X-100), 10 μl dNTP mix (2 mM dATP, dCTP, dGTP, dTTP). After adding water ...
Embodiment 2
[0117] Example 2: Phagemid presentation and screening of hNGAL mutant proteins against human Tear lipocarin.
[0118] 200 ml of a culture containing cells transformed with a plasmid vector similar to phNGAL5 but encoding a library of lipocalin muteins as fusion proteins was transferred into sterile Erlenmeyer flasks. After infection with VCS-M13 helper phage (Stratagene) at a multiplicity of infection of about 10, the culture was then shaken at 160 rpm for 30 minutes at 37°C. Kanamycin (70 μg / ml) was then added, the temperature of the incubator was lowered to 30°C, and anhydrotetracycline (200 μl of a 100 μg / ml stock solution in dimethylformamide (DMF)) was added to 100 μg / L after 10 minutes. to induce gene expression. The incubation was continued for another 5 hours at 30°C, 160 rpm.
[0119] From this culture, 50 ml were taken and the cells were pelleted by centrifugation (15 min, 12000 g, 4°C). The supernatant containing phagemid particles was sterile filtered (0.45 μm...
Embodiment 3
[0128] Embodiment 3: Use " colony screening " method to identify the hNGAL combined with human Tear fat cardin mutein
[0129] In order to produce hNGAL muteins with Strep-Tag(R) II and the albumin binding domain as fusion proteins for analysis and to identify them by colony screening, the gene cassette between the two BstXI cleavage sites was extracted from the vector phNGAL5 Subcloned into phNGAL7.
[0130] For this purpose, plasmid DNA was isolated using the Perfectprep Plasmid Midi Kit (Eppendorf) from the E. coli cloning mixture obtained by infection with phagemids from Example 2 eluted as a result of the final selection round. The DNA was cut with restriction enzyme BstXI, and the smaller of the two fragments (347 bp) was purified by preparative agarose gel electrophoresis as described in Example 1. The DNA of vector phNGAL7 was cut with BstXI and the larger of the two fragments (3971bp) was isolated in the same way.
[0131] For ligation, 100 fmol each of the two ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| pore size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com