Universal primer PCR method for immunocapture to detect bacteria
A technology of universal primers and bacteria, applied in biochemical equipment and methods, measuring devices, and microbial determination/inspection, etc., can solve the problems of difficult to achieve rapid specific detection, complicated operation steps, etc., and achieve clear PCR product bands, Improved sensitivity and high repeatability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0015] Experimental strains: Shigella dysenteriae type I, Shigella sonnei, Shigella flexneri 1a, 2a, 2b, 3a, 4, 5, Y varieties, abalone Shigella boydii 1 . Among them, Shigella dysenteriae type I, Shigella sonneri, Shigella flexneri 4, Shigella baumannii 16 were purchased from the Jiangxi Provincial Health and Epidemic Prevention Station, Shigella flexneri 1a, 2a, 2b, 3a, 5, The Y variant was purchased from the Health and Epidemic Prevention Station of Fujian Province.
[0016] Universal primer: Synthesized by Sangon Company, selected from the conserved region of bacterial 16S rRNA gene, identified as pure by electrophoresis. The primer sequences are as follows: forward primer 5'-AAACTCAAAGGAATTGACGG-3'reverse primer 5'-GACGGGCGGTGTGTACAA-3'.
[0017] Diagnostic serum: The diagnostic serum of the above-mentioned Shigella bacteria is the product of the Lanzhou Institute of Biological Products, Ministry of Health, and the antibodies are purified by the 4-step ammonium sulfate ...
Embodiment 2
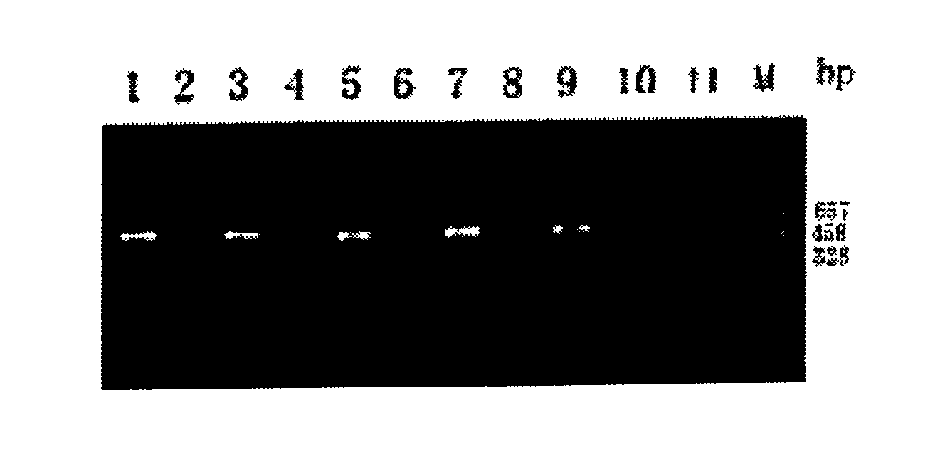
[0030] Neutralization experiment: set up a neutralization group and a control group at the same time to carry out the neutralization experiment. The materials and iUPPCR operation steps are the same as those in Example 1. Dilute Shigella dysenteriae type I, Shigella sonei, Shigella flexneri 4, and Shigella baumannii 16 into 20 μL of bacterial liquid containing 50 CFU of pathogenic bacteria . In the neutralization group, 20 μL of bacterial solution was mixed with an equal volume of 50 μg / mL, 100 μg / mL, and 200 μg / mL of the corresponding antibody, incubated at 37°C for 1 hour, and then added to the reaction plate; for the control group, 20 μL of bacterial solution and 20 μL of sterile water were added. , and the rest of the steps remain unchanged. The neutralization results are shown in Table 1. There was no PCR amplification product in the neutralization group of the three groups, while the corresponding control group had amplification products.
[0031] Table 1: Neutralizati...
Embodiment 3
[0034] Embodiment 3: iUPPCR cross experiment
[0035] 1) Crossover experiments were carried out with purified antibodies from species-specific monovalent diagnostic serum: the materials were the same as in Example 1, with Shigella dysenteriae type I, Shigella sonneri, Shigella flexneri 4 and Shigella baumannii 16 Antibodies from the monovalent diagnostic serum were used to coat reaction plates, and each antibody was coated with 4 wells, which were used to capture 4 kinds of Shigella, and the rest of the steps were the same as in Example 1. The PCR product identification results showed that there was no cross-reaction between species (see Table 2).
[0036]Table 2: Results of the crossover experiment for the detection of Shigella dysenteriae type I, Shigella sonneri, Shigella flexneri 4 and Shigella baumannii 16 using iUPPCR captured by specific monovalent antibodies
[0037] Shigella dysenteriae type I Shigella sonneri Shigella flexneri 4 Shigella baumannii 16 An...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com