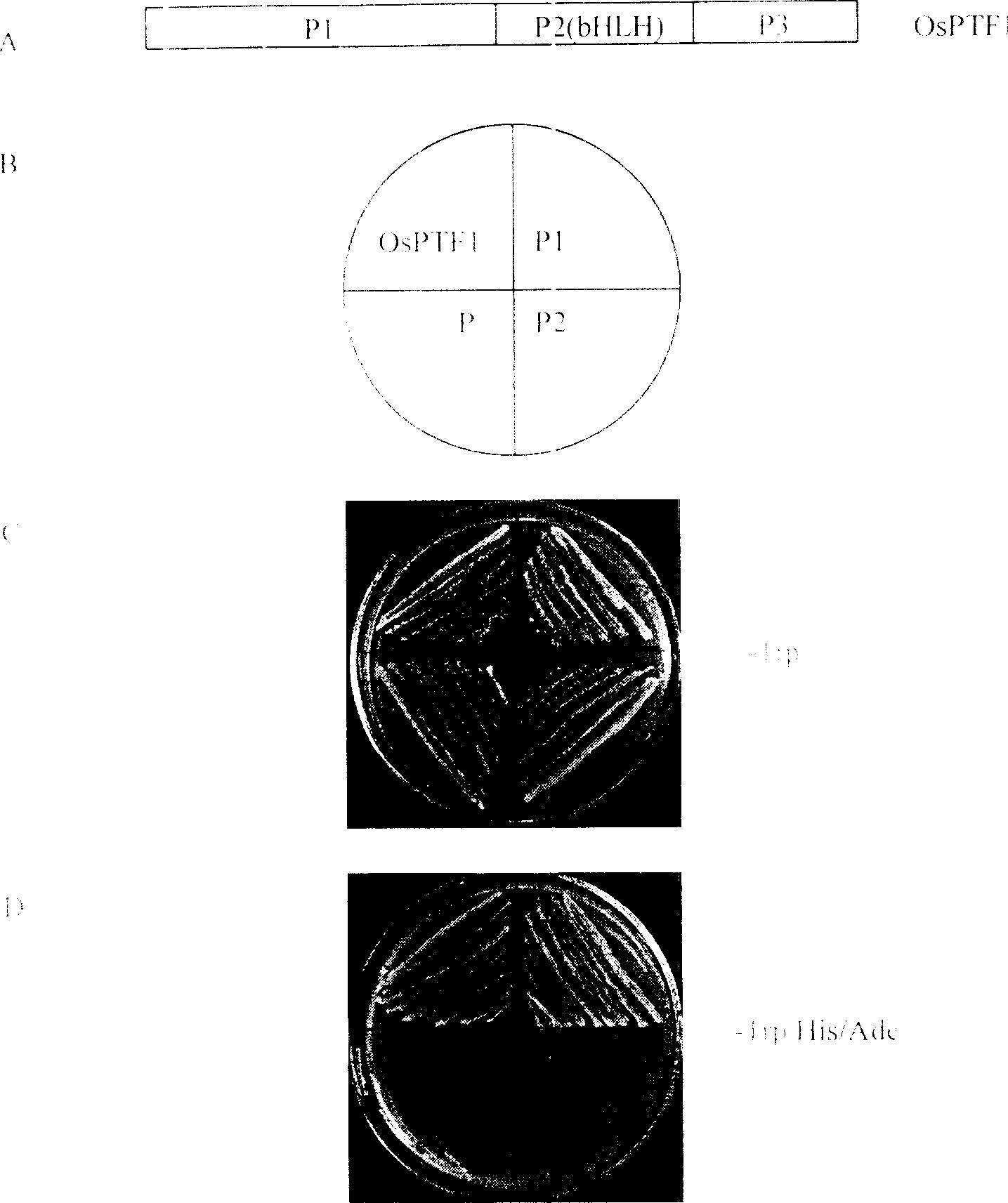
Plant phosphorus hunger inducing transcription factor OsPTF1
A technology of phosphorus starvation and plant application in the field of genetic engineering, which can solve the problems of not being able to be absorbed and utilized by plants, and achieve the effects of improving phosphorus starvation resistance, reducing the application of phosphorus fertilizers, and reducing food production costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] The rice Kasalath variety was soaked at 37°C to accelerate germination, sowed in a yellow sand pot, and transplanted to hydroponic culture after 7 days (the hydroponic culture formula is the standard formula of the International Rice Institute). After the seedlings were cultivated to the three-leaf stage, they began to be divided into groups, one group was cultured normally, and the other group was cultured with phosphorus starvation. After cultivating for 4 days, the materials were collected, and the root materials were collected respectively, and the total RNA was extracted with Trizol from Gibco Company. Oligotex mRNA Mini Kit from Qiagen was used to extract mRNA from 250 g of total RNA, and 1 g of mRNA from each of the two materials was used for cDNA synthesis using SMARTTM PCR cDNA Synthesis Kit from Clontech. The cDNA of normal cultured root tissue was used as Driver, and the cDNA of starved material was used as Tester. The PCR-Select cDNA Subtraction Kit of Clon...
Embodiment 2
[0029] Using the cDNA materials for constructing the suppressive subtraction hybridization library, the PCR-Select Differential Screening kit was used to screen the suppressive subtraction hybridization library to obtain OsPTF1 gene fragment clones. The clone was sequenced and primers designed for rapid amplification of cDNA ends (RACE). Primer 1: ATTTCACAAGGGCCAACAGACAT, for 3' end RACE amplification, primer 2: CCCCCATATTTTTCCGATTTC, for 5' end RACE amplification. RACE amplification uses SMART RACE cDNA Amplification Kit from Clontech. The PCR product was cloned into the pT-Adv vector using the PCR Cloning Kit of Clontech Company. After transformation, the plasmid was extracted for sequencing. Finally, the sequence was spliced using computer software to obtain a full-length cDNA sequence, a total of 1862bp. The detailed sequence is shown in SEQ ID No.1, wherein the open reading frame is located at nucleotides 67-1503. The amino acid sequence of OsPTF1 was deduced accordin...
Embodiment 3
[0030] The rice variety Kasalath that was normally cultured to the three-leaf stage began to be stressed by phosphorus starvation, and the treatment method was consistent with the process of constructing a suppressive subtraction hybrid library. After 4 days of stress, the roots and leaves of normal seedlings and the roots and leaves of starvation-treated seedlings were taken respectively. Total RNA was extracted with Trizol reagent from Gibco. 5 g of total RNA was taken from each of the 4 materials for reverse transcription. The reverse transcription process is as follows: add 5 μg RNA to a 1.5 μl centrifuge tube, 1 μl Oilgo(dT) 15 (Promega Company), add RNase-free water to 8 μl, denature in a 70°C water bath for 5 minutes, cool on ice for 5 minutes, add 4 μl 5×First Strand Buffer (Invitrogen Company), 2 μl 0.1DTT (Invitrogen Company), 4 μl 2.5mMdNTPs (Takara Company), 1 μl Rasin (40unit / l) (Promega Company) and 1 μl SuperScript RTII (Invitrogen Company), mixed with a gun, ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com