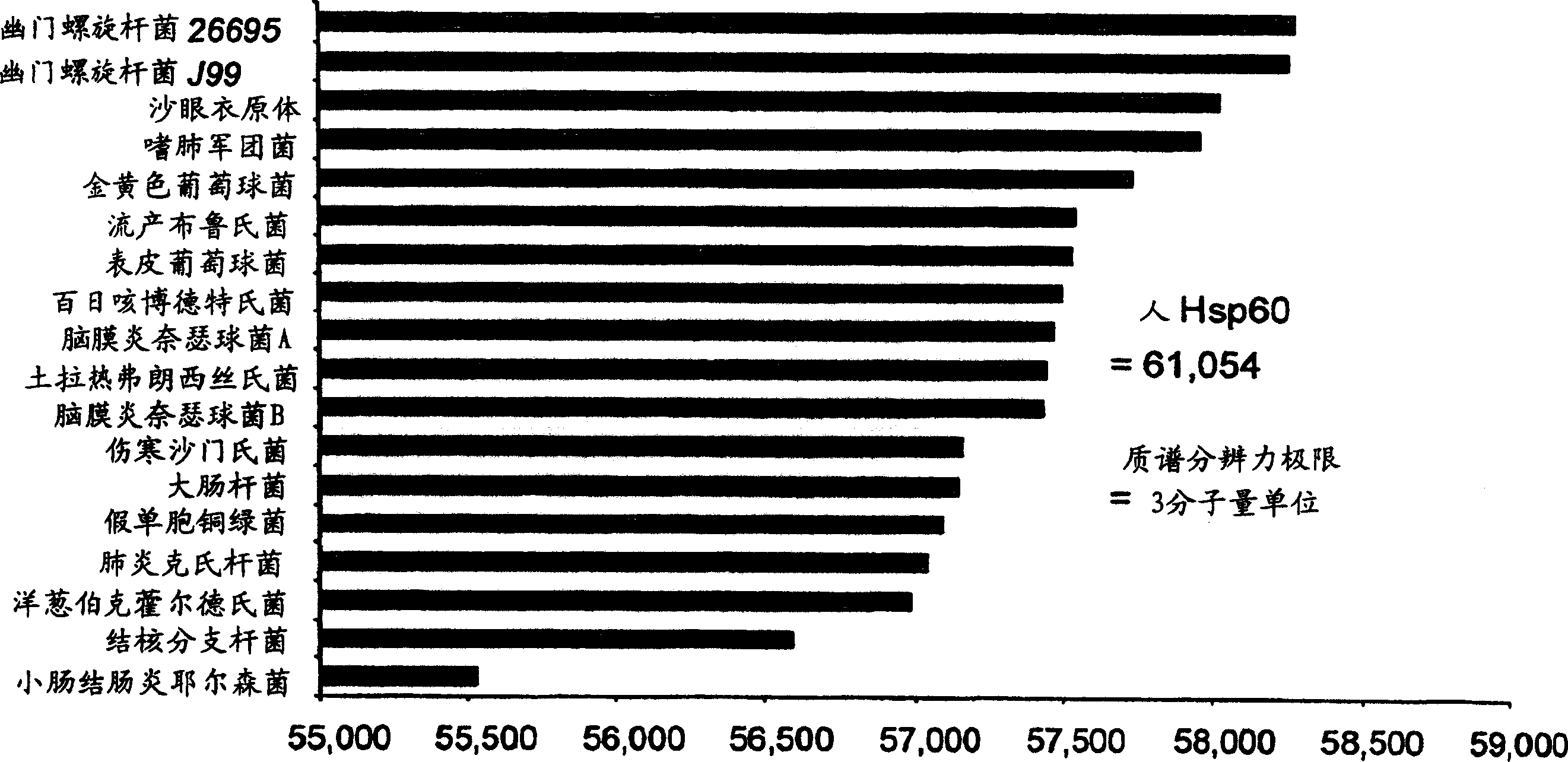Identifying micro-organisms
A microbial and biomarker technology, applied in biological testing, microbial determination/inspection, biological material analysis, etc., can solve the problem of not recommending affinity purification and the use of biomarkers
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
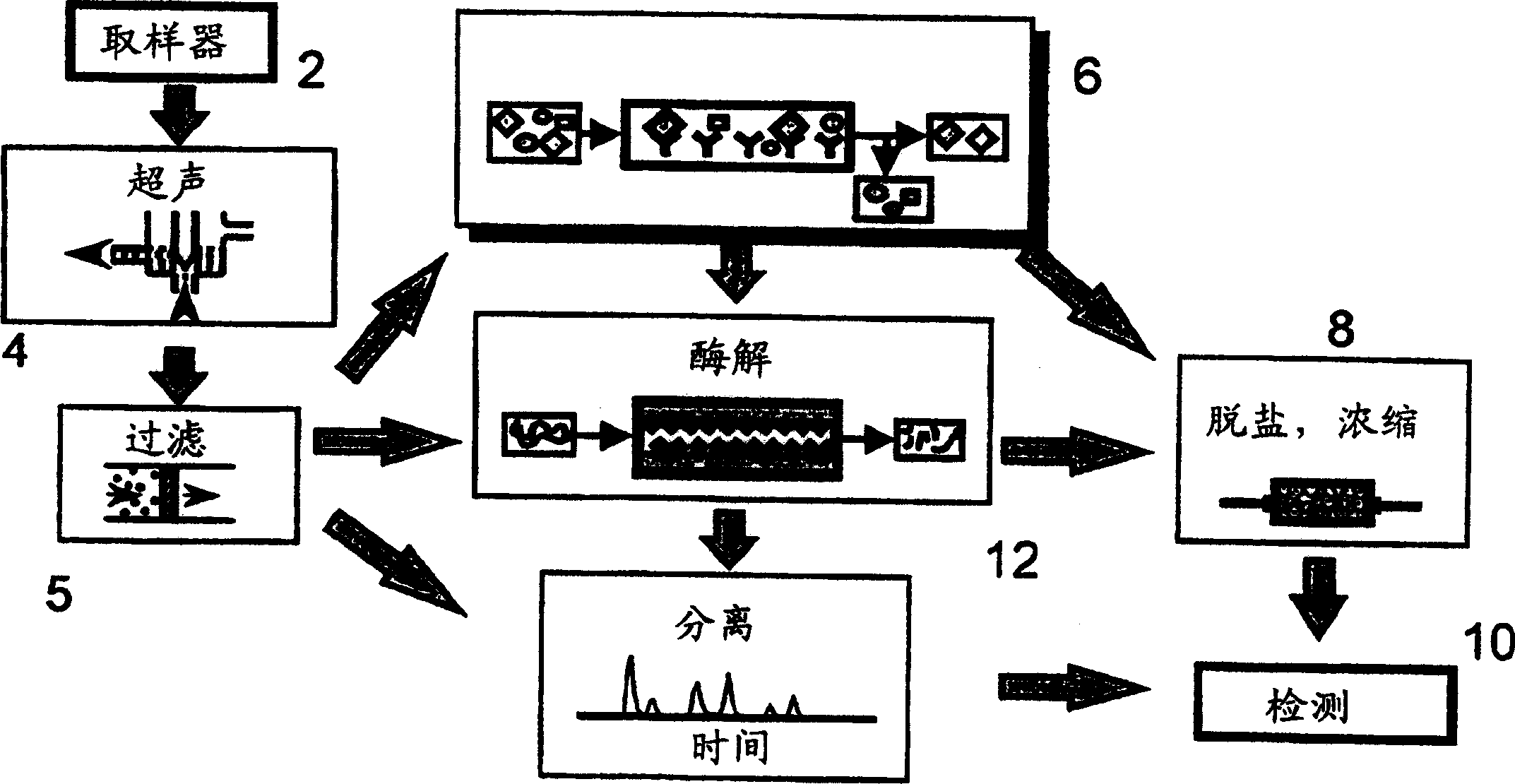
[0056] Example 1 Automatic sampling and identification system
[0057] Referring to Figure 1, a vacuum device (not shown) is used to obtain a sample from an aerosol suspected of containing pathogenic bacteria. The aerosol is mixed with the carrier liquid, and the suspension is fed into the system (1) through the sampler (2). From there, the suspension is transported to the ultrasonic sound generator (4), in which the cell wall of any bacteria in the suspension is broken up with ultrasound, thereby releasing the constituent proteins of the bacteria into the lysis solution. Inevitably, the lysate also contains fragments. Therefore, downstream of the ultrasonic sound generator (4) is a filter (5), and the filter (5) prevents unwanted substances from passing through. In some cases, the use of detergents can promote lysis, which should not affect the downstream immunoaffinity step. Suitable mild non-ionic detergents are well known in the art, including polyoxyethylene-based detergents ...
Embodiment 2
[0061] Example 2 Using Hsp60 as a biomarker to identify potential pathogens
[0062] The average molecular weight of Hsp60 obtained from a variety of organisms can be predicted with high accuracy from the known amino acid sequence (corrected based on the presence of isotopic mixtures) or directly measured with a suitable purified recombinant protein. Although Hsp60 is highly conserved in many species (not only bacteria), mass spectrometry can determine the molecular weight with a high degree of accuracy, and can distinguish protein molecules with only three molecular weight units. Comparing these measured values with the known value database can identify the relevant species, as shown in Table 1.
[0063] Hsp60M r
58015.3Da
Chlamydia trachomatis
57301.7Da
57154.8Da
Salmonella typhimurium
56757.3Da
Burkholderia pseudomallei
[0064] Figure 2 shows the molecular weight of Hsp60 in a va...
Embodiment 4
[0148] Example 4 Comparison of Arg-CHsp60 peptides of Brucella of the genus Brucella and Staphylococcus epidermidis
[0149] The endopeptidase Arg-C (Rhizobacter protease), as its name implies, cleaves the carboxy-peptide bond of arginine. Figure 4 is the peptide fingerprint obtained by Arg-C digestion of Brucella and Staphylococcus epidermidis Hsp60 Icon Compare. As shown in Figure 3, the molecular weights of intact Hsp60 proteins from these organisms are similar (57649 and 57529, respectively, including N-terminal methionine). However, the peptide set obtained is very different and characterizes related organisms.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com