Probe and method for detecting heterosigma akashiwo
A technology for the detection of Akashio algae and detection probes, which is applied in the field of molecular biology detection of microorganisms in the marine environment, can solve the problems of lack of research and achieve good species specificity and small sample volume
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0016] Example 1. Preparation of nucleotide sequences for detection of Akashiwa algae
[0017] The algae used in the present invention are isolated from natural seawater samples of Dalian Bay. Pure cultures are obtained by repeatedly picking individual algal cells under a microscope. The culture medium used is conventional F / 2 culture medium, culture conditions: light / dark cycle 12h / 12h, light intensity 4000 Lx, culture temperature 22°C-25°C.
[0018] Use a 0.45 μm microporous membrane or centrifuge at 10,000 r / min to collect the algae cells in the logarithmic growth phase of Akashiwa algae, and use TE buffer (10mmol / L Tris-HCl (Dihydroxymethylaminomethane hydrochloric acid) pH8.0 , 1mmol / LEDTA (ethylenediaminetetraacetic acid) pH8.0) to wash about 20mg of algae, add 2 times the volume of extraction buffer (3% (w / v) CTAB (hexadecane base-trimethyl-ammonium bromide), 1% (w / v) sarkosyl (sodium lauryl sarcosinate), 20mmol / L EDTA, 1.4mol / L NaCl, 0.1mol / LTris-HCl pH8.0 , 1% (v / v...
Embodiment 2
[0020] Design and synthesis of embodiment 2.H.akashiwo primer1 and H.akashiwo primer2
[0021] By comparing with the sequences of the corresponding regions of all known other eukaryotes and prokaryotes in Genbank, it is found that the sequence shown in SEQ ID NO.1 is very different from the corresponding sequences of other organisms. Based on the sequence, two nucleic acid molecular probes of H.akashiwo primer1 and H.akashiwo primer2 were designed. According to the nucleotide composition and arrangement of the two probes or their complementary sequences, the nucleotide sequences described in SEQ ID NO.2 and SEQ ID NO.3 can be obtained on a commercial DNA synthesizer.
Embodiment 3
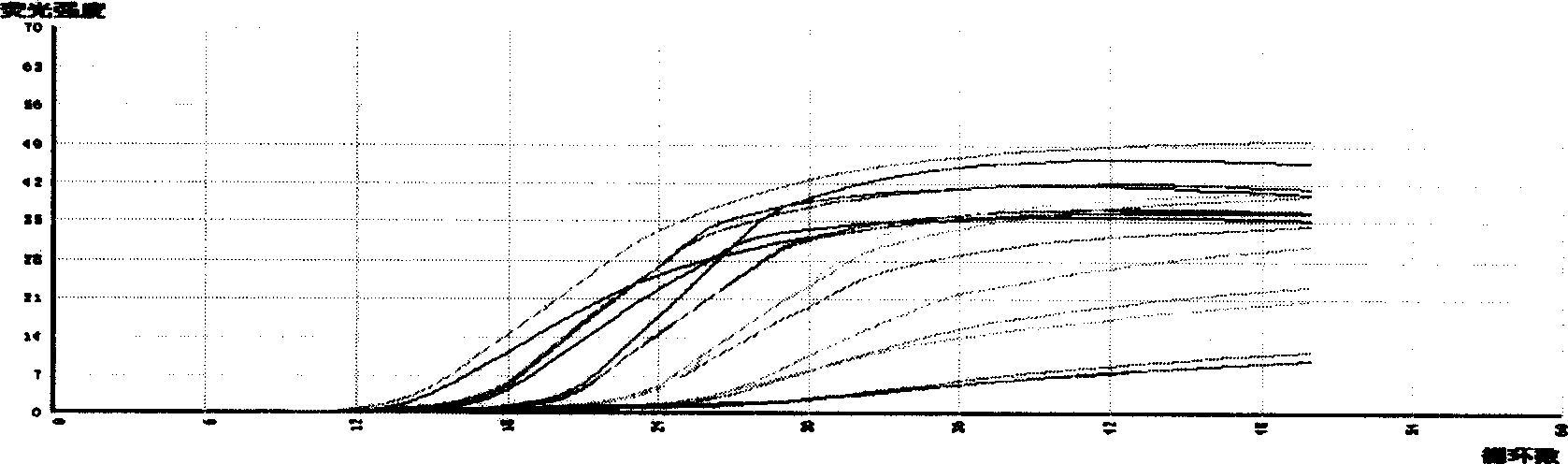
[0022] Example 3. Detection of Akashiwa algae by conventional PCR method
[0023] In this embodiment, several strains of microalgae in the algae bank of this laboratory were selected as reference algae, and they were: Chaetoceros curvisetus (Chaetoceros curvisetus), Chaetoceros curvisetus, C. C. gracile, C. minimum, Gymnodinium sp., G. mikimotoi, Thalassiosira nordenskioeldii, Pseudonitzschia pungens, Nitzschia closterium, Navicula membranacea, Melosira sp., Skeletonemacostatum, isolated from Tamar in Mirs Bay Alexandrium tamarens, they all obtain pure cultures by repeatedly picking single algal cells under a microscope. These algae were cultured and the DNAs of these algae and I. akashiwo algae were extracted according to the method described in Example 1.
[0024] Get 1 μ l of DNA extracts of various algae, carry out PCR according to the PCR reaction system described in Example 1, with the nucleus described in SEQ ID NO.2 (H.akashiwo primer1) and SEQ ID NO.3 (H.akashiwo pr...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com