Fumor invasion and metastasis resisting function and use of venin cysteine proteinase inhibitor
A cysteine protease and inhibitor technology, applied in the field of medical biology, can solve the problem that snake venom cysteine protease inhibitors have not been reported in literature and the like
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] Example 1: Cloning of sv-Cystatin cDNA
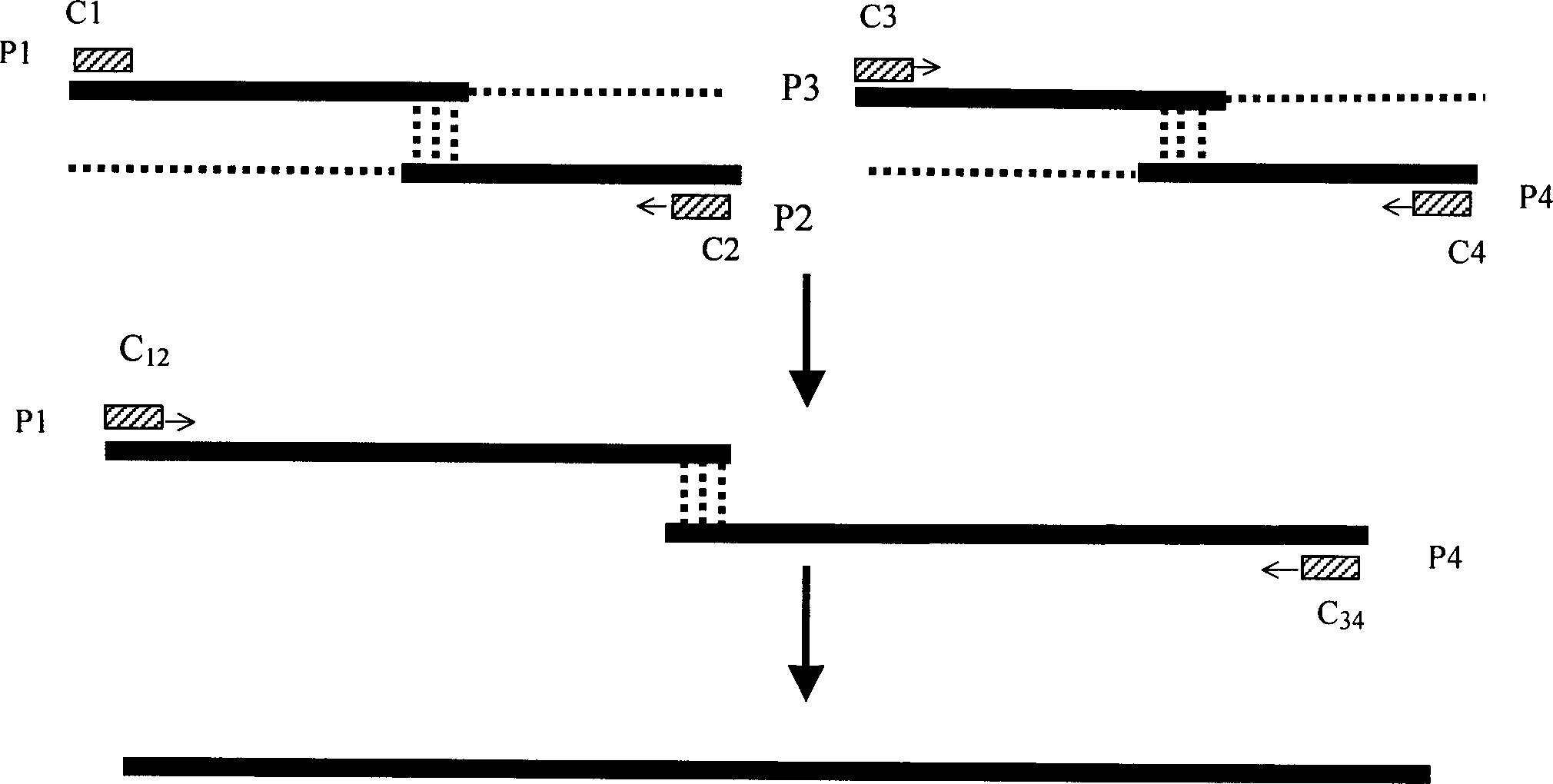
[0045] According to the 99 amino acid sequence (SEQ ID NO.1) of the Chinese cobra venom cysteine protease inhibitor (sv-Cystatin) protein, according to the yeast dominant codon, deduce its cDNA sequence (SEQ ID NO.2); according to SEQ ID NO.2 sequence, design four partially complementary oligonucleotides C1, C2, C3 and C4 corresponding to artificial synthesis, and 4 pairs of PCR primers P1, P2, P3 and P4.
[0046] Its sequence is as follows:
[0047] C1 sequence:
[0048] ATCCCAGGTGGTTTGTCTCCAAGATCTGTTTCTGACCCAGACGTTC 46
[0049] AAAAGGCTGCIGCIITCGCTGTTCAAGAAATACAACGCTGGTTCTG 91
[0050] C2 sequence
[0051] ACTTTTCACCAGCAACAGATTGAGATTGAGCTTCAACAACTTCTCAA 46
[0052] TTCCTTGTAGTAGTGAGCGTTAGCAGAACCAGCGTTGTATTCTTG 91
[0053] C3 sequence
[0054] ATCTGTTGCTGGTGAAAAGTACTTCTTGATGATGGAATTGGTTAAG 46
[0055] ACTAAGTGTGCTAAGACTGCTGGTAAGCCAAAGGTTTACAAGGA 90
[0056] C4 sequence
[0057] CCAAACTTGGAAACCCACAACTTTTCTTCTTGTTGCTTG...
Embodiment 2
[0064] Example 2: Construction of pPICZ-sv-Cystatin Pichia secretory expression vector
[0065] Using sv-Cystatin cDNA as a PCR template, the upstream primer F1 (5'-CG GAATTC ATCCCAGGTGGTTTGTCTCC-3', the underline is EcoR I restriction site) and downstream primer R1 (5'-GC TCTAGA AACCAAACTTGGAAACCAC-3', the underline is the Xba I restriction site) PCR amplification of the sv-Cystatin DNA fragment containing the corresponding restriction site.
[0066] ECOR I / Xba I double digested sv-Cystatin DNA fragment and Pichia pastoris expression vector pPICZαA, ligated and transformed E.coli.Top10, screened positive recombinants, extracted plasmids for DNA sequencing and identification. The results showed that the sv-Cystatin DNA insert sequence and The reading frame was completely correct, and the constructed vector was named pPICZ-sv-Cystatin.
Embodiment 3
[0067] Example 3: Screening Pichia genetically engineered bacteria that stably and efficiently express sv-Cystatin
[0068] The pPICZ-sv-Cystatin vector was digested with SacI to make it linear, and the Pichia GS115 competent cells prepared by electroporation were transformed with 1M sorbitol, and the transformed bacteria solution was coated on the YPDS plate (YPD Culture medium: 1% yeast extract, 2% peptone and 2% glucose), cultured upside down at 30°C, observe the formation of recombinant colonies after 2-4 days.
[0069] Use a sterilized toothpick to plant the transformed single colony on the MMH and MDH plates (MMH medium: 1.34% yeast medium YNB, 4 × 10 -5 % biotin, 0.5% methanol, 4×10-4 histidine MDH medium: 1.34% YNB, 4×10 -5 % biotin, 2% glucose, 4×10-4 histidine), cultured upside down at 30°C for 2-3 days.
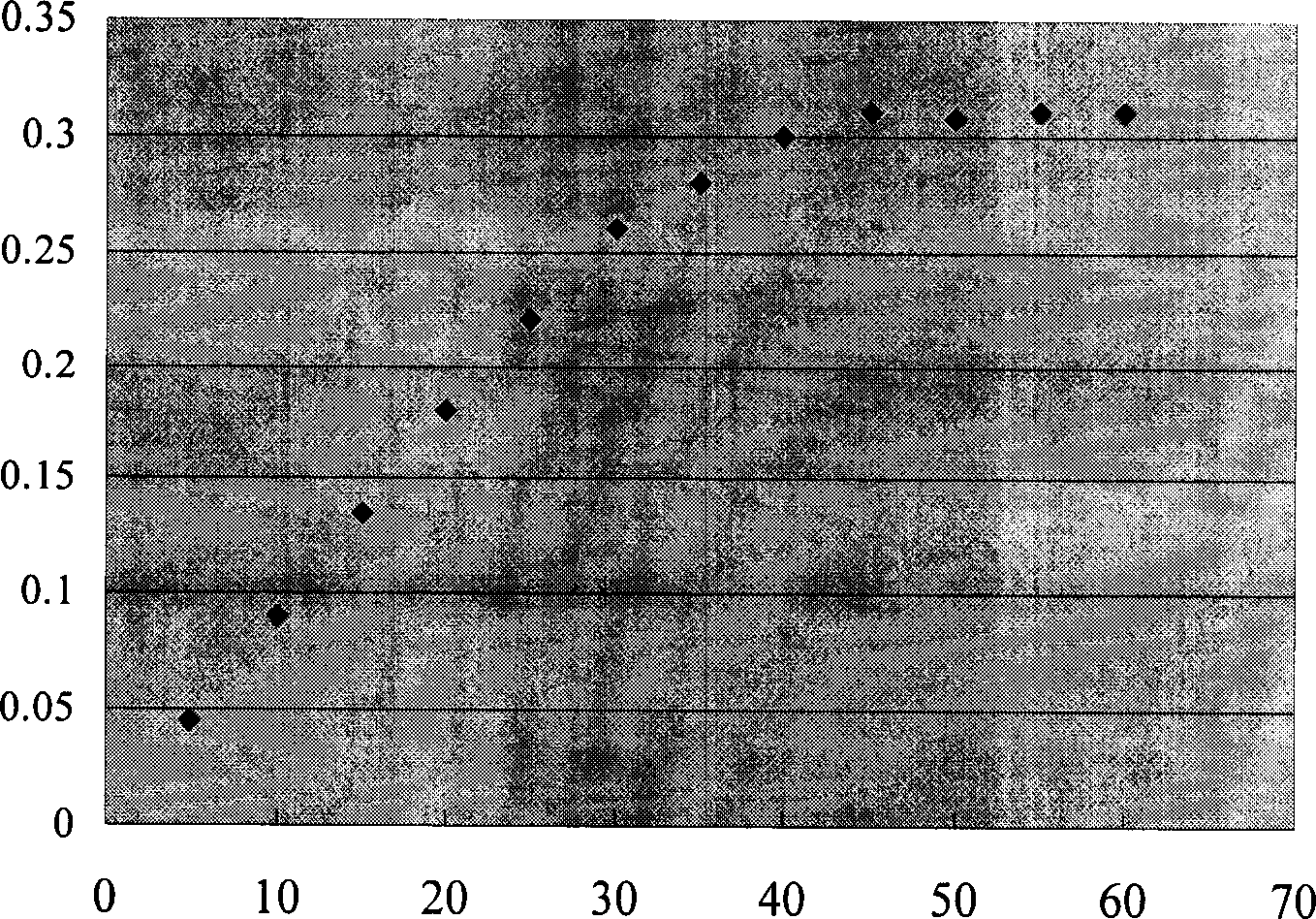
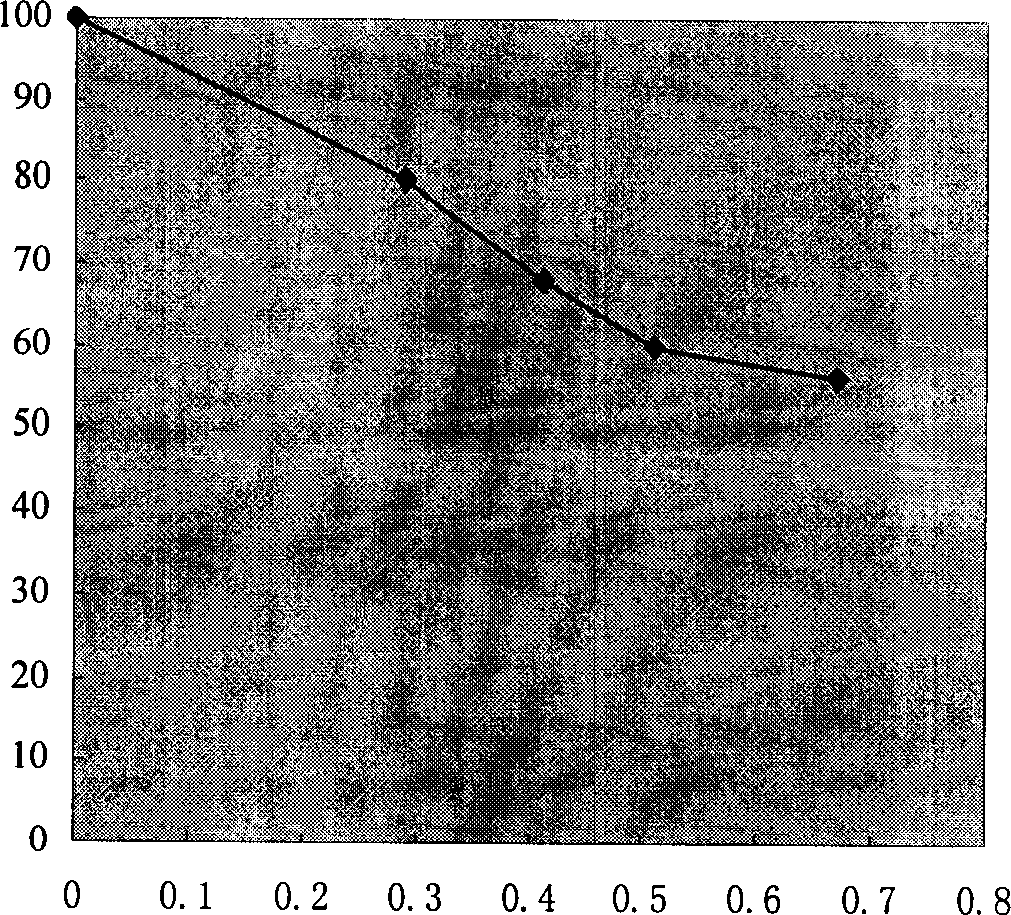
[0070]Judging the phenotype according to the difference in the growth rate of the colony on the MMH and MDH plates: the colony that grows normally on both the MM...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com