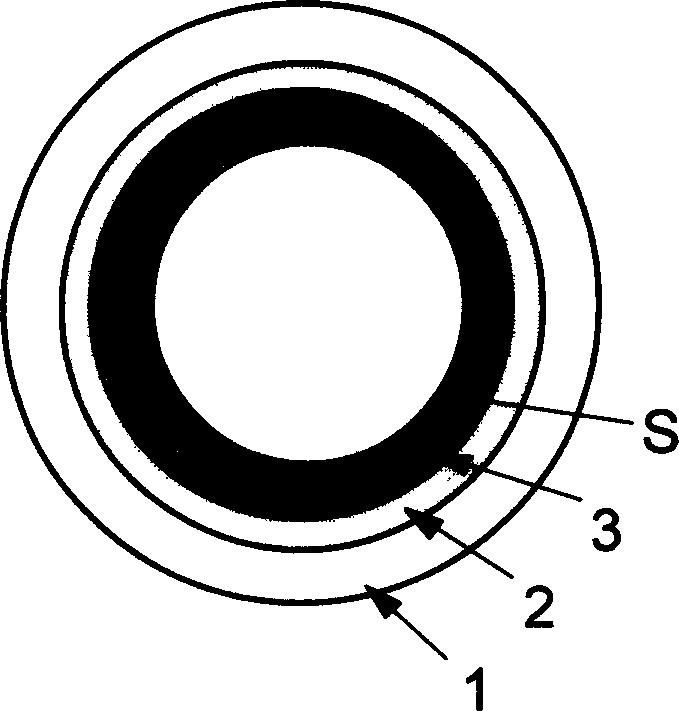
DNA fluorescent capillary biosensor and preparing process thereof
A biosensor and DNA probe technology, applied in the fields of biochemistry and molecular biology analysis, can solve the problems of small DNA probes, complex detection instruments, weak fluorescence signals, etc. economic effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] In this example, the synthesized 100 μmol / L oligonucleotide sequence 5'-GAAA CCTG TTTG TTGG ATAC-3' was used as a DNA probe, which was immobilized on the inner surface of a capillary according to the above-mentioned method to prepare a DNA-FCBS.
[0032] During the measurement, 10 μL of 33 μmol / L target nucleotide sequence 5′-GTATCC AACAAACA GGTTTC-3′ solution was inhaled with DNA-FCBS, and allowed to hybridize at 45°C for 90 minutes; EB) staining; wash out the free EB staining solution in DNA-FCBS with 0.2% SDS and diethyl ester pyrocarbonic acid (DEPC) water; use RF-5000 fluorescence spectrometer to measure at excitation wavelength 526nm, emission wavelength 584nm, the result obtained Yes: the average value of fluorescence intensity before and after hybridization is 74.01±14.12, the statistical analysis result is t=4.1106, p<0.05, and there is a significant difference before and after hybridization.
Embodiment 2
[0034] In this example, 10 μL of 1 μmol / L Cy5-labeled target nucleotide sequence 5′-GTATCCAACAAACA GGTTTC-3 solution was sucked into the DNA-FCBS of the present invention, and hybridized at 45° C. for 90 minutes; washed out with 0.2% SDS and DEPC water Free target nucleotides in DNA-FCBS; measured with RF-5000 fluorescence photometer at excitation wavelength 646nm, emission wavelength 664nm, the results are shown in Table 1; the average value of fluorescence intensity before and after hybridization is 61.66 ± 18.22, statistics The analysis result was t=6.7684, 0.005<P<0.01, and there was a highly significant difference before and after hybridization.
[0035] DNA-FCBS serial number
Embodiment 3
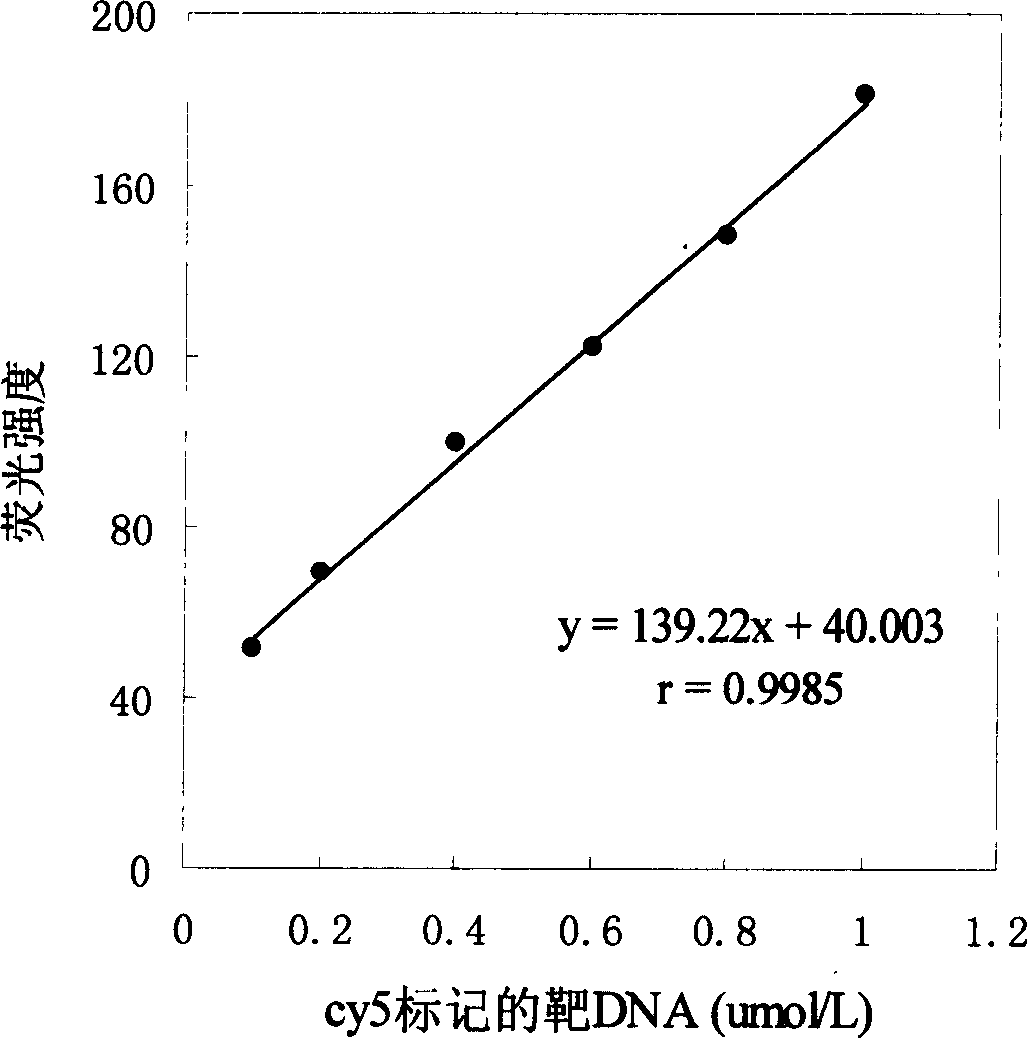
[0037] In this example, under the experimental conditions that the hybridization time is 90min and the hybridization temperature is 45°C, a series of Cy5-labeled target nucleotide solutions with different concentrations are inhaled with the DNA-FCBS of the present invention, and the RF-5000 fluorescence photometer is used to detect 646nm, measured at the emission wavelength of 664nm, the results are as follows figure 2 shown. The Cy5-labeled target nucleotide concentration has a good linear relationship with the fluorescence intensity in the range of 0.1-1 μmol / L: y=139.73x+39.613, the correlation coefficient is 0.9985, and the detection limit is 1.7 pmol.
PUM
| Property | Measurement | Unit |
|---|---|---|
| length | aaaaa | aaaaa |
| thickness | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com